Plant Name: Gentiana manshurica
Taxonomic Information:
Plant ID: NPO20836
Plant Latin Name: Gentiana manshurica
Taxonomy Genus: Gentiana
Taxonomy Family: Gentianaceae
Plant External Links:
NCBI TaxonomyDB:
683755
Plant-of-the-World-Online:
n.a.
Used in Medicines:
Country/Region:
ChinaTraditional Medicine System:
TCMMedicinal Functions:
Antibacterial; Stomachic
Geographical Distribution:
China
Collective Molecular Activities of the Plant
List of Human Proteins Collectively Targeted by the Plant
Click Gene ID to Access Activity Profile of Individual Target
| GPR35; | |
| NPSR1; | |
| RECQL; TDP1; ALOX12; AKR1B1; HSD17B2; HSD17B10; HPGD; NOX4; APEX1; POLB; | |
| TEK; INSR; MET; AXL; KDR; FLT3; CDK1; EGFR; PIM1; SRC; IGF1R; AURKB; CSNK2A1; NUAK1; | |
| CA12; CA7; | |
| TYR; | |
| AHR; | |
| SLC22A6; | |
| SMAD3; HSPA1A; |
List of Human Cytochrome P450 Enzymes Affected by the Plant
Click Gene ID to Access Activity Profile of Individual Enzyme
Detailed Information of Target Proteins
| Protein Class | Gene ID | Protein Name | Uniprot ID | Target ChEMBL ID |
|---|---|---|---|---|
| Cytochrome P450 family 1 | CYP1A2 | Cytochrome P450 1A2 | P05177 | CHEMBL3356 |
| Cytochrome P450 family 1 | CYP1B1 | Cytochrome P450 1B1 | Q16678 | CHEMBL4878 |
| Cytochrome P450 family 1 | CYP1A1 | Cytochrome P450 1A1 | P04798 | CHEMBL2231 |
| Enzyme_unclassified | ALOX12 | Arachidonate 12-lipoxygenase | P18054 | CHEMBL3687 |
| Enzyme_unclassified | AKR1B1 | Aldose reductase | P15121 | CHEMBL1900 |
| Enzyme_unclassified | HSD17B2 | Estradiol 17-beta-dehydrogenase 2 | P37059 | CHEMBL2789 |
| Enzyme_unclassified | HSD17B10 | Endoplasmic reticulum-associated amyloid beta-peptide-binding protein | Q99714 | CHEMBL4159 |
| Enzyme_unclassified | HPGD | 15-hydroxyprostaglandin dehydrogenase [NAD+] | P15428 | CHEMBL1293255 |
| Enzyme_unclassified | NOX4 | NADPH oxidase 4 | Q9NPH5 | CHEMBL1250375 |
| Enzyme_unclassified | APEX1 | DNA-(apurinic or apyrimidinic site) lyase | P27695 | CHEMBL5619 |
Showing 1 to 10 of 36 entries
Enrichment of Gene Ontology of Target Proteins (Activity≤1μM)
Detailed Information of Enriched Gene Ontolgy
| GO Type | GO Category | Enriched GO Terms | p-Value | Adjusted p-Value | Enriched Genes |
|---|---|---|---|---|---|
| BP | GO:0009987; cellular process | GO:0043066; negative regulation of apoptotic process | 1.574E-10 | 1.359E-07 | AKR1B1, ALOX12, AURKB, AXL, CDK1, CSNK2A1, EGFR, HSPA1A, IGF1R, KDR, PIM1, SMAD3, SRC, TEK |
| BP | GO:0009987; cellular process | GO:0051897; positive regulation of protein kinase B signaling | 1.121E-08 | 4.354E-06 | AXL, EGFR, INSR, MET, NOX4, SRC, TEK |
| BP | GO:0008152; metabolic process | GO:0038083; peptidyl-tyrosine autophosphorylation | 2.441E-08 | 8.305E-06 | EGFR, IGF1R, INSR, KDR, SRC |
| BP | GO:0009987; cellular process | GO:0070301; cellular response to hydrogen peroxide | 9.291E-08 | 2.273E-05 | AKR1B1, APEX1, AXL, CDK1, CYP1B1 |
| BP | GO:0032502; developmental process | GO:0031100; animal organ regeneration | 2.828E-07 | 6.051E-05 | AXL, CDK1, CSNK2A1, EGFR, FLT3 |
| BP | GO:0008152; metabolic process | GO:0055114; oxidation-reduction process | 4.252E-07 | 8.818E-05 | AKR1B1, ALOX12, APEX1, CYP1A1, CYP1A2, CYP1B1, HPGD, HSD17B10, HSD17B2, NOX4, TYR |
| BP | GO:0008152; metabolic process | GO:0097267; omega-hydroxylase P450 pathway | 7.070E-07 | 1.400E-04 | CYP1A1, CYP1A2, CYP1B1 |
| BP | GO:0008152; metabolic process | GO:0045740; positive regulation of DNA replication | 3.238E-06 | 5.424E-04 | CDK1, EGFR, IGF1R, INSR |
| BP | GO:0009987; cellular process | GO:0006286; base-excision repair, base-free sugar-phosphate removal | 3.377E-06 | 5.529E-04 | APEX1, POLB |
| BP | GO:0032502; developmental process | GO:0007568; aging | 3.955E-06 | 6.286E-04 | APEX1, AURKB, CDK1, CYP1A1, NOX4, POLB |
Showing 1 to 10 of 39 entries
Enriched KEGG Pathways of Target Proteins (Activity≤1μM)
Top 10 KEGG Pathways (ordered by adjusted p-value)
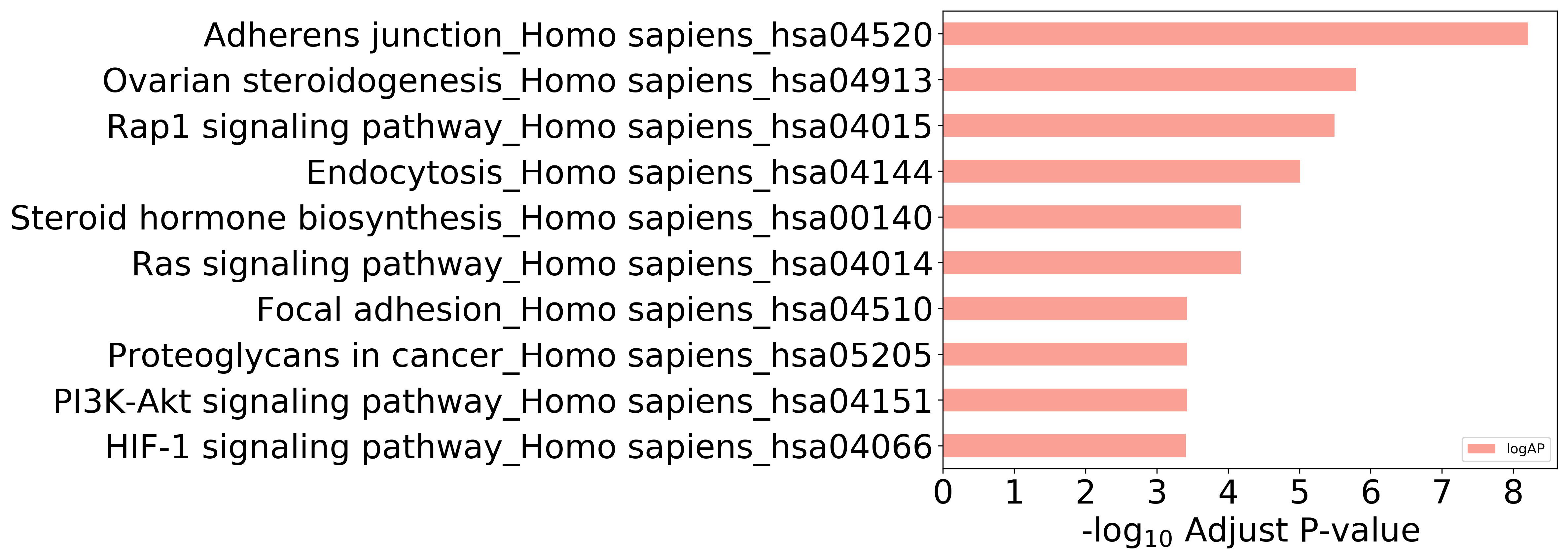
Detailed Information of Enriched KEGG Pathways
| Pathway Category Top Level | Pathway Category Second Level | Pathway ID | Pathway Name | p-Value | Adjusted p-Value | Enriched Genes |
|---|---|---|---|---|---|---|
| 09100 Metabolism | 09103 Lipid metabolism | hsa00140 | Steroid hormone biosynthesis | 3.499E-06 | 6.649E-05 | CYP1A2, HSD17B2, CYP1A1, CYP1B1 |
| 09100 Metabolism | 09105 Amino acid metabolism | hsa00380 | Tryptophan metabolism | 5.055E-05 | 5.239E-04 | CYP1A2, CYP1A1, CYP1B1 |
| 09100 Metabolism | 09111 Xenobiotics biodegradation and metabolism | hsa00980 | Metabolism of xenobiotics by cytochrome P450 | 3.055E-04 | 1.935E-03 | CYP1A2, CYP1A1, CYP1B1 |
| 09100 Metabolism | 09102 Energy metabolism | hsa00910 | Nitrogen metabolism | 4.212E-04 | 2.452E-03 | CA12, CA7 |
| 09120 Genetic Information Processing | 09124 Replication and repair | hsa03410 | Base excision repair | 1.606E-03 | 7.323E-03 | POLB, APEX1 |
| 09130 Environmental Information Processing | 09132 Signal transduction | hsa04015 | Rap1 signaling pathway | 8.479E-08 | 3.222E-06 | SRC, INSR, KDR, TEK, MET, EGFR, IGF1R |
| 09130 Environmental Information Processing | 09132 Signal transduction | hsa04014 | Ras signaling pathway | 2.932E-06 | 6.649E-05 | INSR, KDR, TEK, MET, EGFR, IGF1R |
| 09130 Environmental Information Processing | 09132 Signal transduction | hsa04151 | PI3K-Akt signaling pathway | 2.974E-05 | 3.791E-04 | INSR, KDR, TEK, MET, EGFR, IGF1R |
| 09130 Environmental Information Processing | 09132 Signal transduction | hsa04066 | HIF-1 signaling pathway | 3.443E-05 | 3.924E-04 | INSR, TEK, EGFR, IGF1R |
| 09130 Environmental Information Processing | 09132 Signal transduction | hsa04068 | FoxO signaling pathway | 9.334E-05 | 8.868E-04 | SMAD3, INSR, EGFR, IGF1R |
Showing 1 to 10 of 26 entries
Human Diseases Associated with the Target Proteins of the Plant
Detailed Information of Human Diseases
| ICD10 Disease Category | Disease Name | ICD10 Code | Associated Disease Targets (Association Ref: TTD database) |
|---|---|---|---|
| A00-B99: Certain infectious and parasitic diseases | Cryptosporidium infection | A07.2 | EGFR; |
| A00-B99: Certain infectious and parasitic diseases | Human immunodeficiency virus infection | B20-B26 | FLT3; |
| C00-D49: Neoplasms | Acute lymphoblastic leukemia | C91.0 | FLT3; |
| C00-D49: Neoplasms | Acute myeloid leukemia | C92.0 | AURKB; FLT3; |
| C00-D49: Neoplasms | Hematological malignancies | C81-C86 | AURKB; SRC; |
| C00-D49: Neoplasms | Metastatic breast cancer | C50 | KDR; |
| C00-D49: Neoplasms | Glioma | C71 | KDR; EGFR; |
| C00-D49: Neoplasms | Head and neck cancer | C07-C14, C32-C33 | KDR; EGFR; AKR1B1; |
| C00-D49: Neoplasms | Advanced cancers | C00-C96 | EGFR; |
| C00-D49: Neoplasms | Advanced renal cell carcinoma | C64 | KDR; |
Showing 1 to 10 of 114 entries


