Collective Molecular Activities of Useful Plants
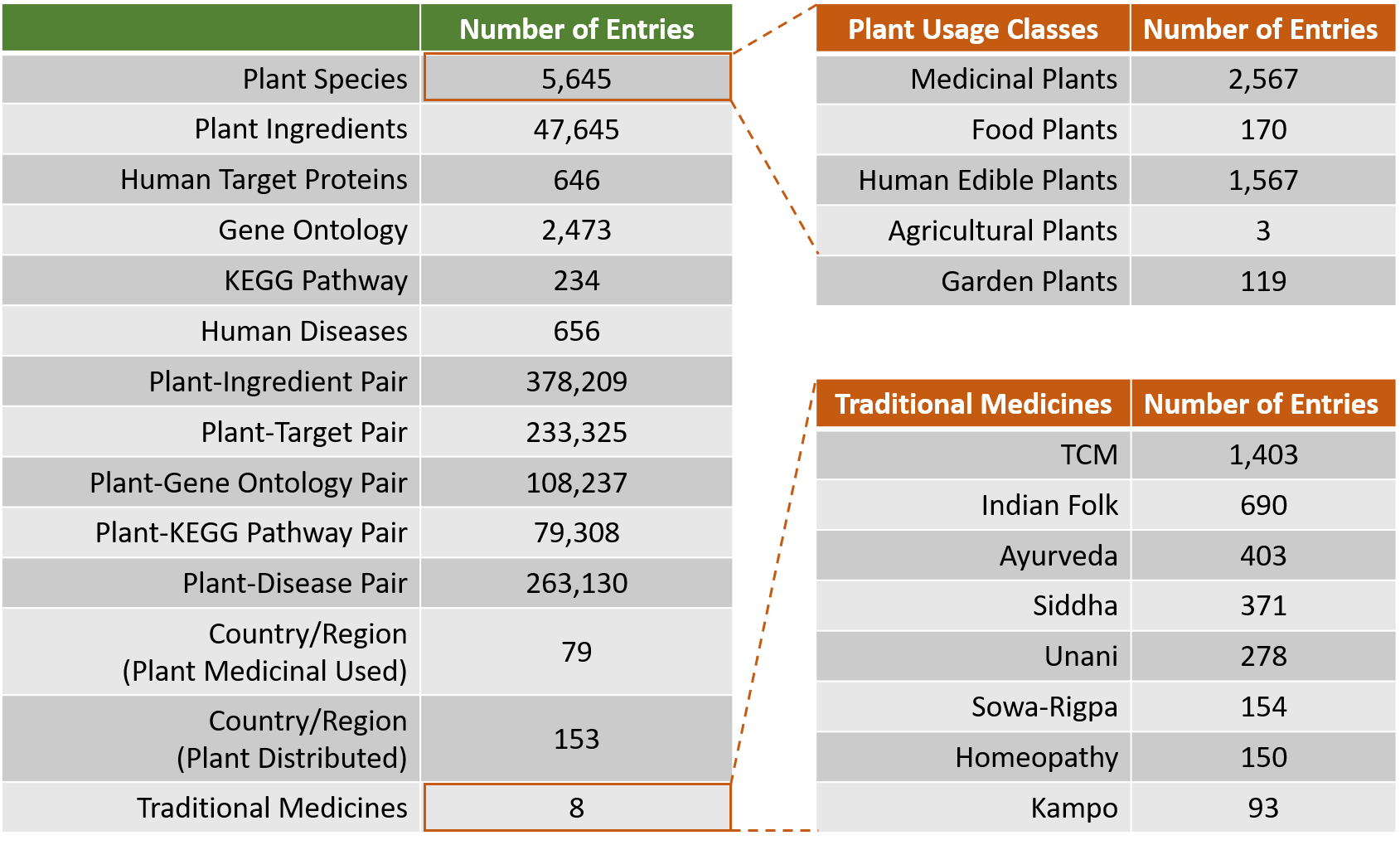
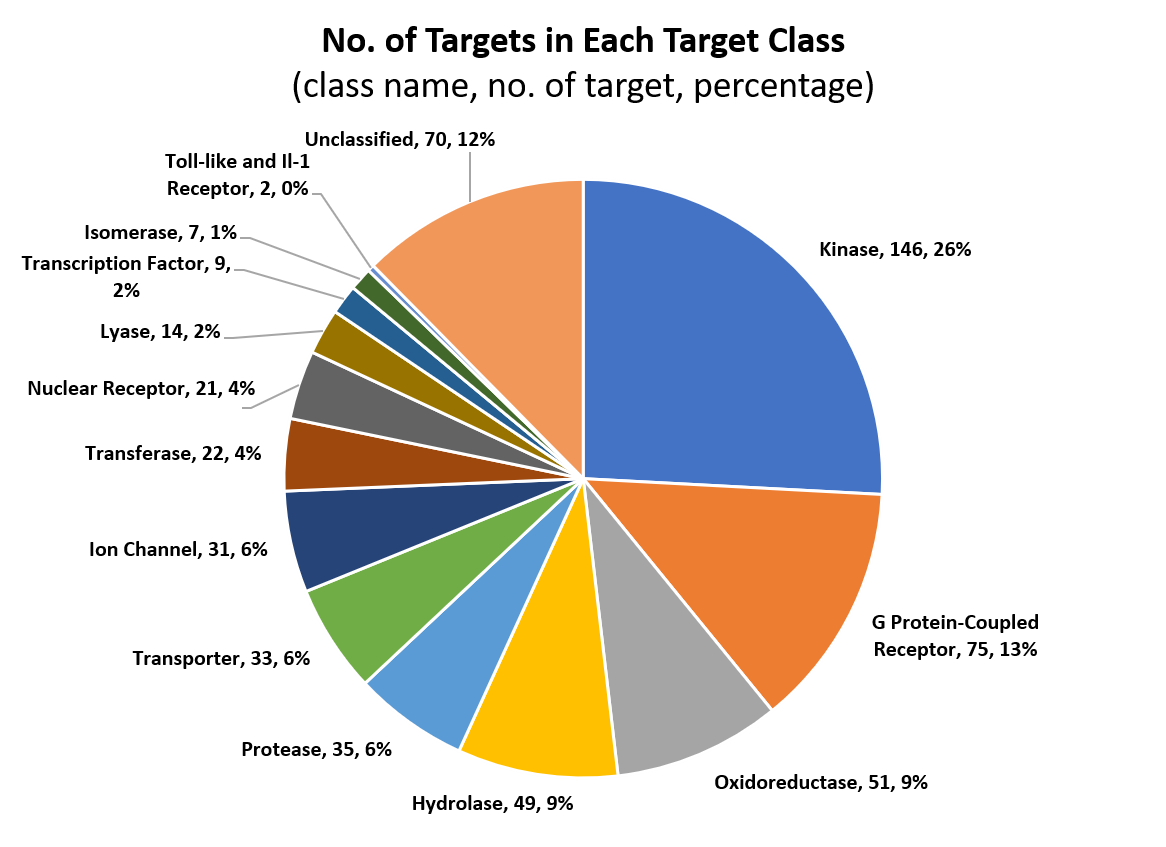
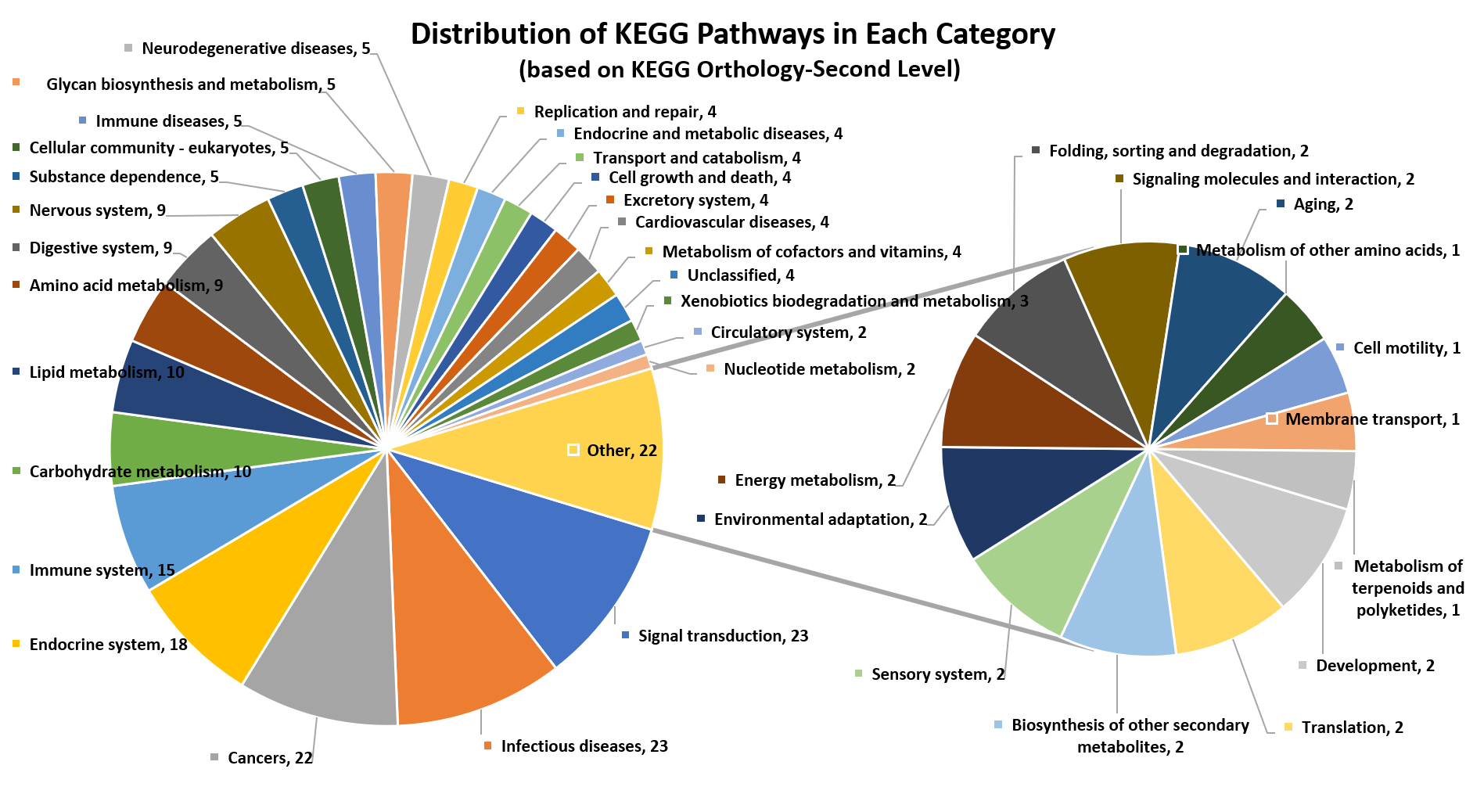
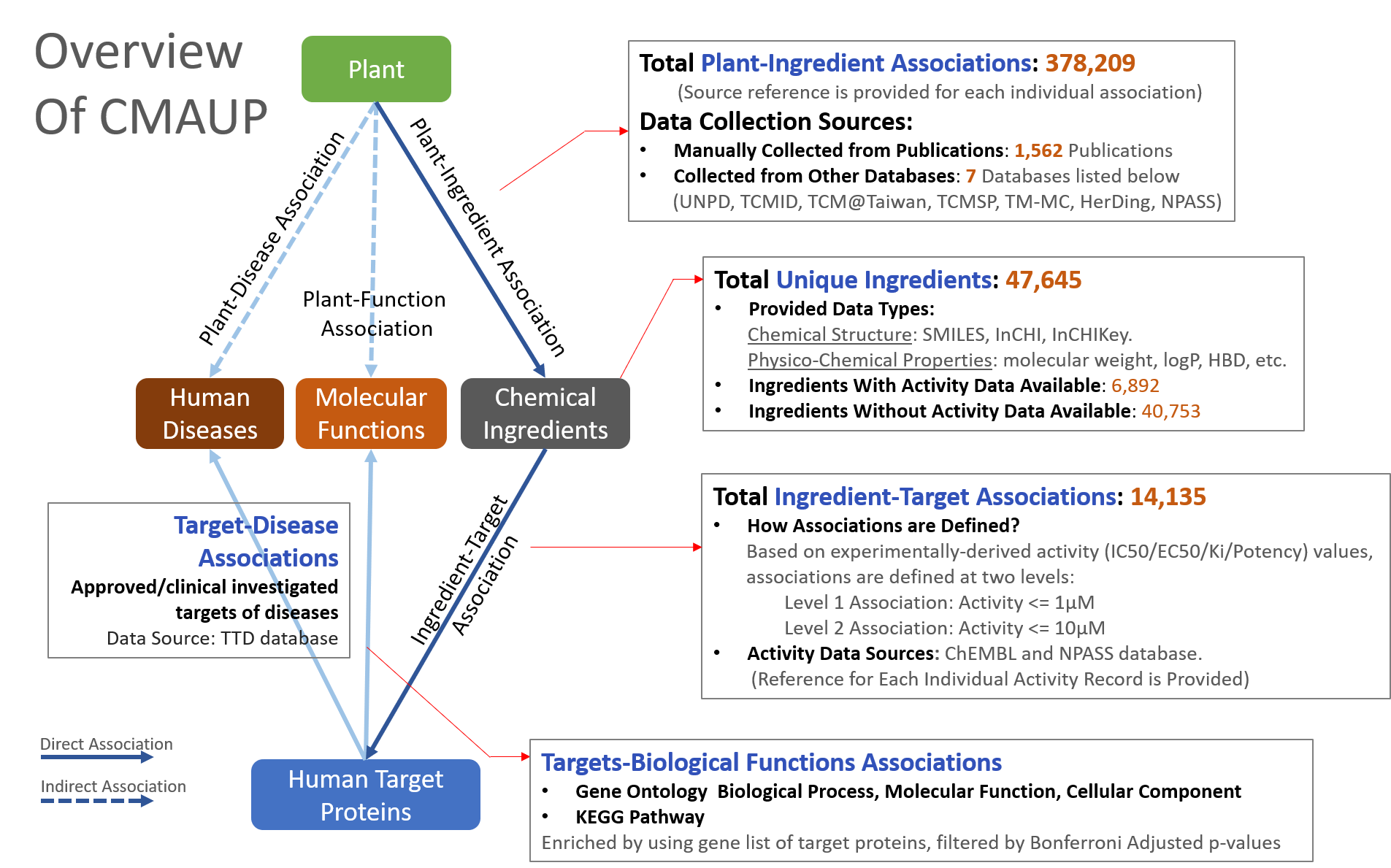
The CMAUP database provides collective molecular activities of 5,645 useful plants, including 2,567 medicinal plants used in 79 countries/regions, on 646 human target proteins and 2,473 Gene Ontology, 234 KEGG pathways, and their relations to 656 human diseases.
To develop the database, we extensively curated chemical ingredients of plants either by manually inspecting 1,562 literatures or from other related databases, resulting in a comprehensive Plant-Ingredient Association dataset. Next, biological activities of these ingredients were collected from ChEMBL and NPASS database by matching ingredient's chemical structure. Plant medicinal usage information was manually collected from literatures or from few traditional medicine databases.
Only activities that ingredients against human target proteins with activity type of IC50/EC50/AC50/Ki/Potency and activity unit of nM were included for further analysis (activities on non-human proteins were excluded). After annotating activities of ingredients of individual plants, proteins that with activities ≤1000nM (≤10000nM was used as level-2 cutoff) were defined as human target proteins of the plant. Gene list of the human target proteins were subjected to Gene Ontology and KEGG pathway enrichment analysis.
Approved or clinical investigated therapeutical targets and their associated human diseases were collected from Therapeutical Target Database (TTD). A human disease was linked to plants when one or more target proteins of the plants are target(s) of the disease, based on TTD data.
Features
>> Multiple Search/Browse Options
- Search/Browse Plants by Plant Usage Class
- Search/Browse Plants by Target Protein
- Search/Browse Plants by Target Gene Ontology
- Search/Browse Plants by Target KEGG Pathway
- Search/Browse Plants by Related Disease
- Browse Plants by Geographical Location
- Search Plants by Chemical Ingredient
>> Medicinal Plants Used in Various 26 Asian, 19 African, 23 European, 8 North and South American, and 3 Oceania Countries/Regions
>> Collective Molecular Activities of 5,645 Plants Based on Experiment-Derived (not in-silico predicted) Quantitative Activities
>> View Detailed Activity Values Along With Source References
>> Free Download of All Data
External Links to Other Resources
Entries in CMAUP database were linked to various external databases, including:
- Plant databases: NCBI TaxonomyDB, Plants of teh World Online
- Chemical Databases: PubChem, ZINC, ChEMBL
- Activity and Therapeutical related databases: Uniprot, KEGG, QuickGO, ChEMBL, TTD
- Literature Resources: PubMed, doi.org

Statistics of Data Content