1. BROWSE THE DATABASE
By clicking each section under the “Browse CMAUP Database”, users will be redirected to a new page where one can browse the database by multiple ways. Alternatively, users can click the button "Browse" on the top of the homepage to browse plants.
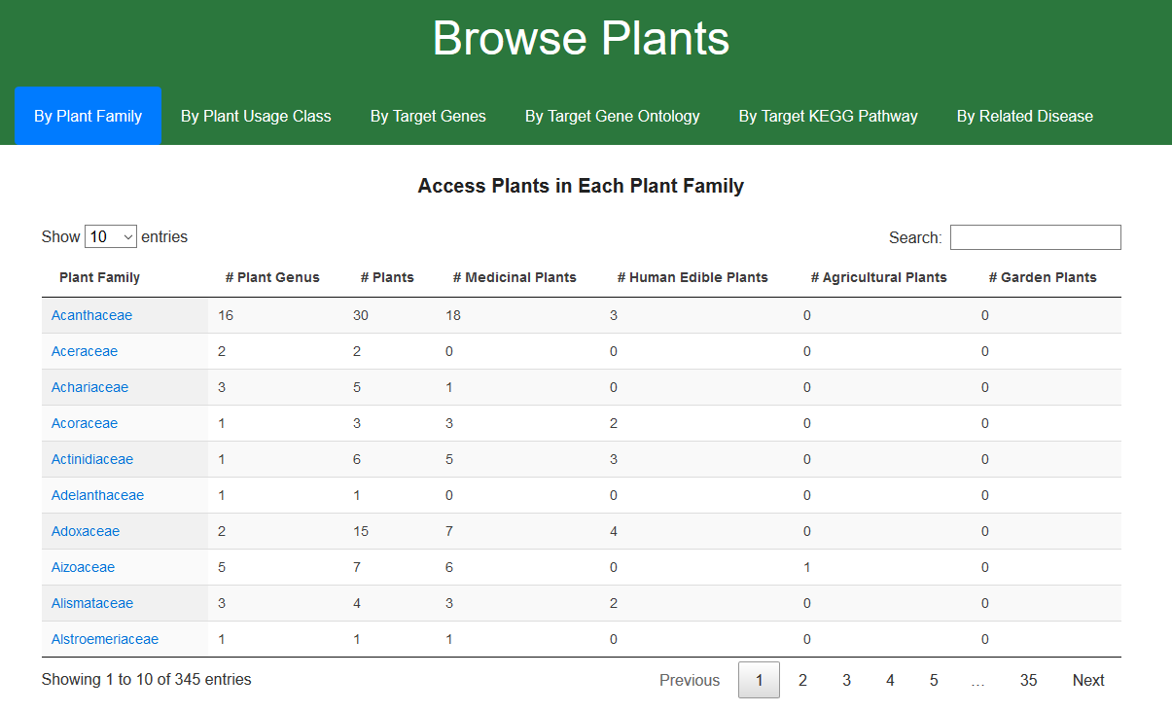
1.1 Browse CMAUP by plant family
The current version of CMAUP includes 5754 plant species which from 345 taxonomic families. Users can access plants of a specific family by click the family name in the first column of the table.
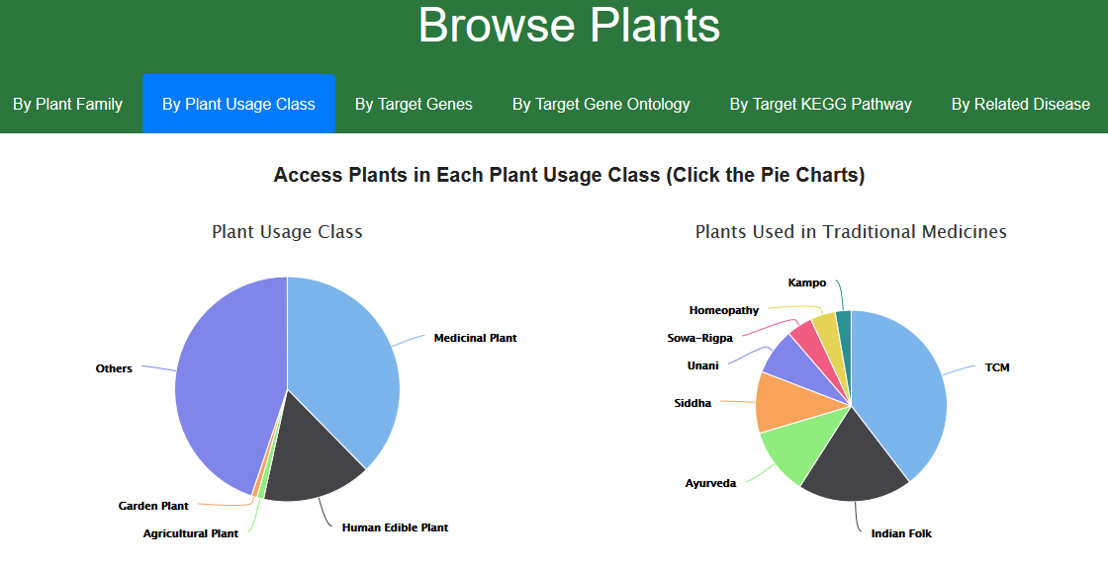
1.2. Browse CMAUP by plant usage class
Generally, plants are categorized into several usage classes: Medicinal Used Plant; Human Edible Plant; Agricultural Plant; Garden Plant; and Others. Moreover, the medicinal used plants are further labelled with traditional medicine systems that uses the plant as medicinal herb. These traditional medicine systems include: TCM (Traditional Chinese Medicine); Indian Folk; Ayurveda; Siddha; Unani; Sowa-Rigpa; Homeopathy; and Kampo. Users can access plants of each category by clicking the pie charts.
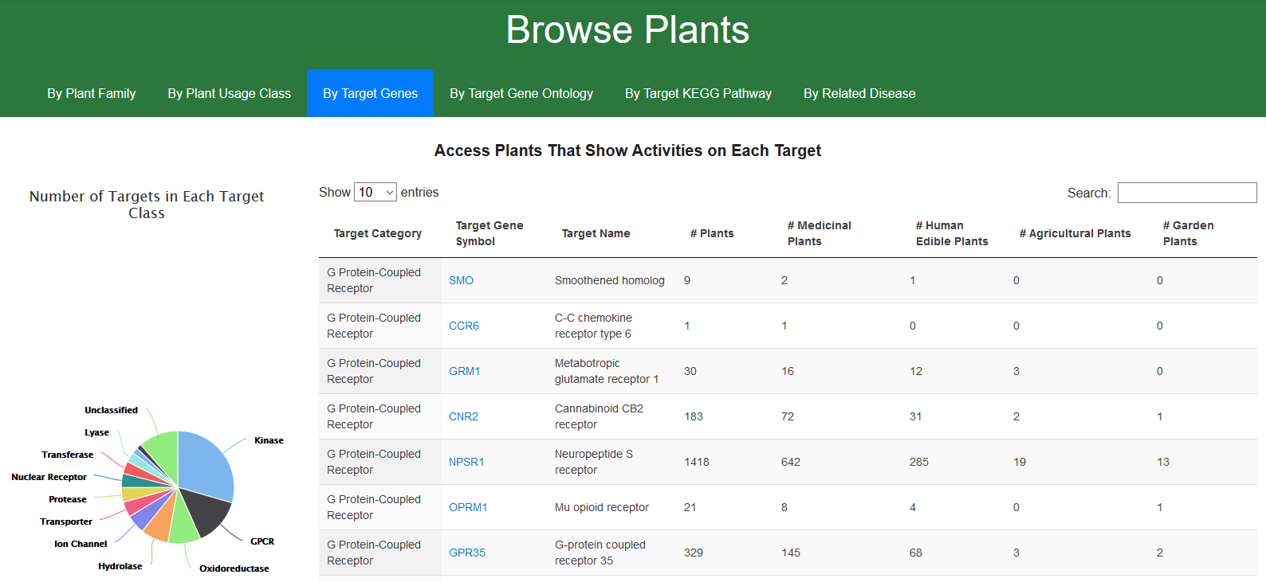
1.3. Browse CMAUP by target proteins
Target proteins of plants are classified into few categories: Kinase; Oxidoreductase; Protease; GPCR; Ion Channel; Transporter; Nuclear Receptor; Unclassified and so on. Users can re-order the targets by each table columns such as “target Category”, then access plants by a specific target (click the target gene symbol).
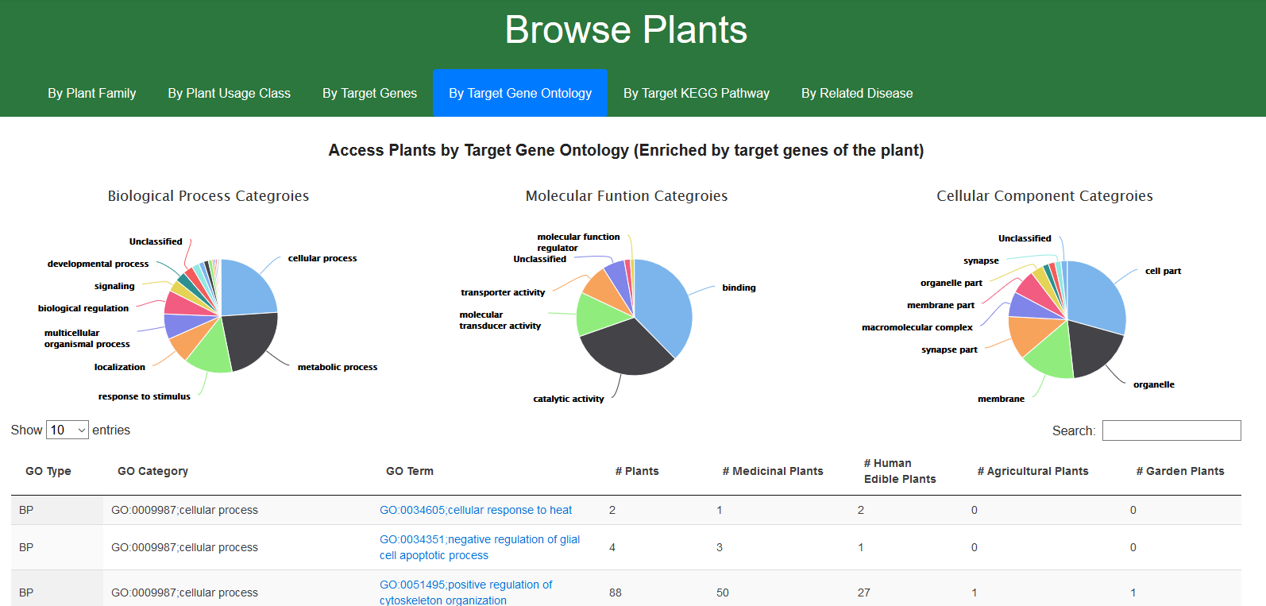
1.4. Browse CMAUP by target gene ontology
Gene ontology (GO) terms were enriched for genes of protein targets of each plant. GO terms are categorized by the categories of GO hierarchical system. Users can re-order the GO terms by their categories and access plants of a specific GO term by clicking the “GO Term”.
1.5. Browse CMAUP by target KEGG pathway
Genes of protein targets of plants were also enriched for KEGG pathways. Users can access plants of a specific pathway by clicking the pathway name.

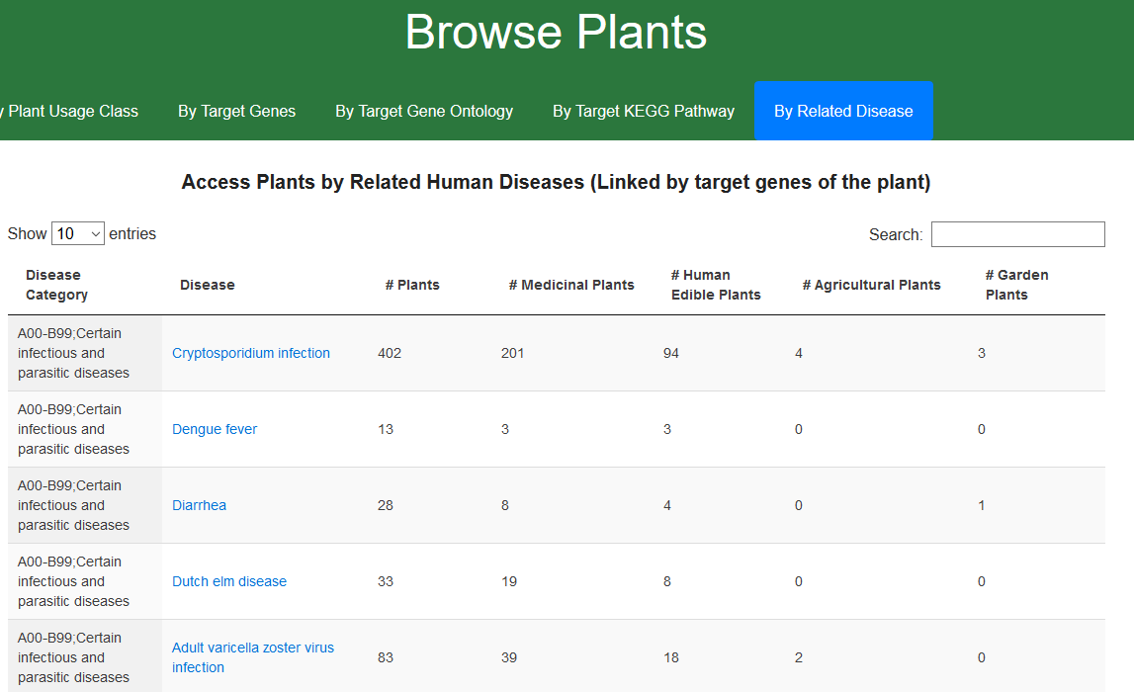
1.6. Browse CMAUP by related human disease
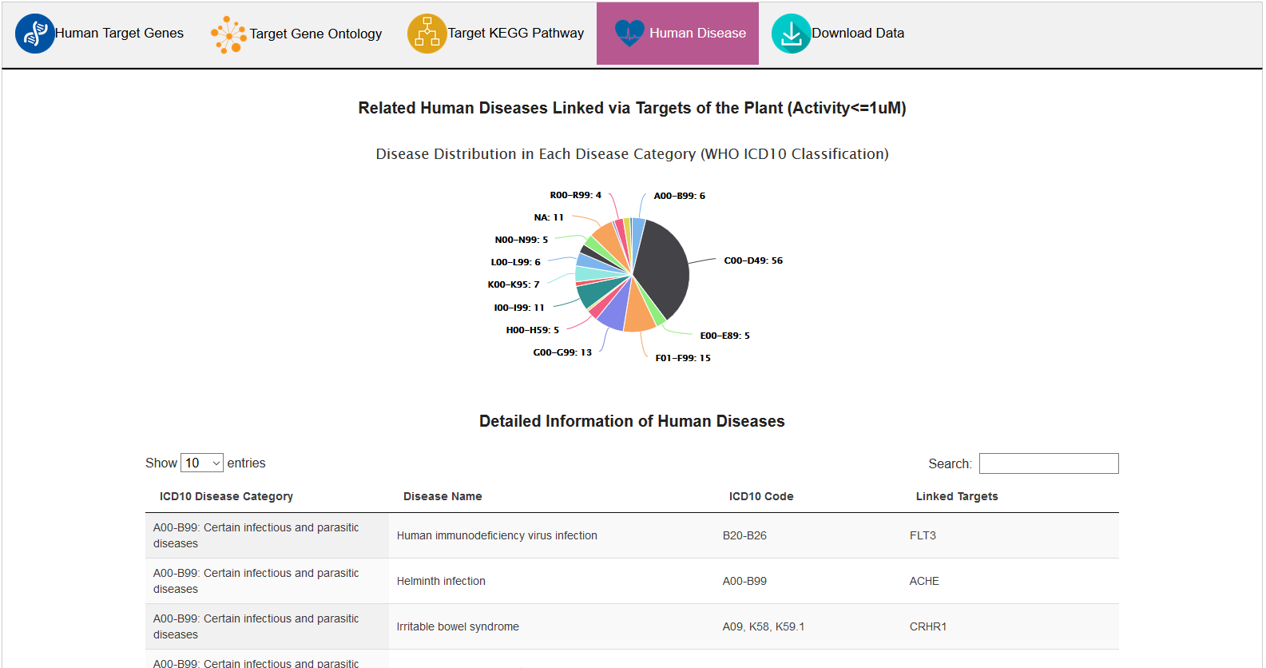
Plants are linked to human diseases via the targets. Diseases are categorized by the ICD-10 system. Users can access plants of a related human disease by clicking the disease name.
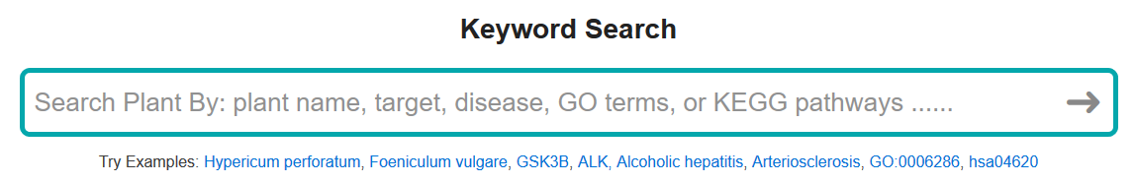
2. SEARCH BY KEYWORDS
Users can search plants by multiple type of keywords: plant name, target gene symbol/protein name, disease, GO term ID/name, and KEGG pathway.
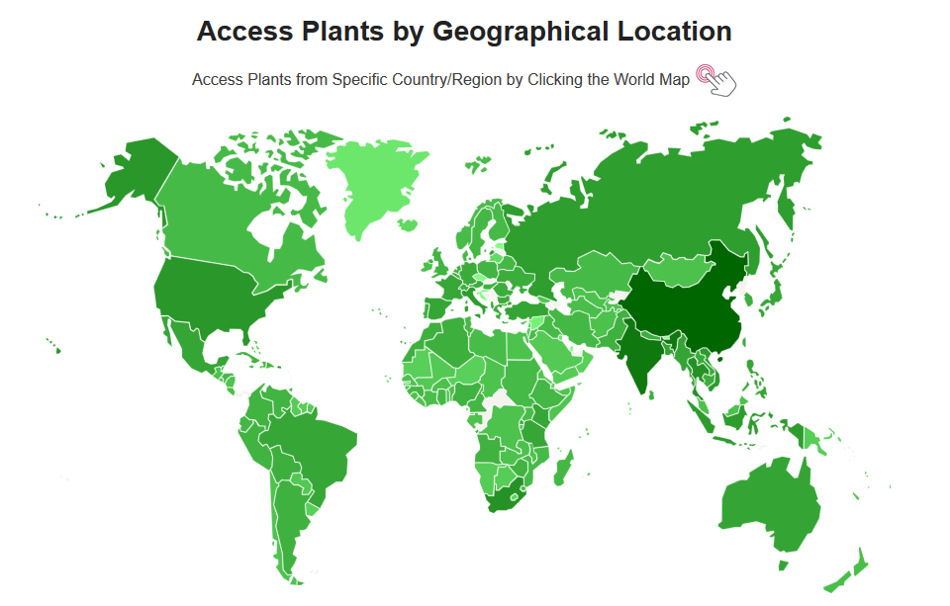
3. ACCESS PLANTS BY WORLD MAP
Users can click country/region on the world map to access related plants.
4. DOWNLOAD DATA
All data can be freely downloaded at each individual webpage.

5. WEBPAGE OF A SPECIFIC PLANT
A typical example of plant page is shown below:
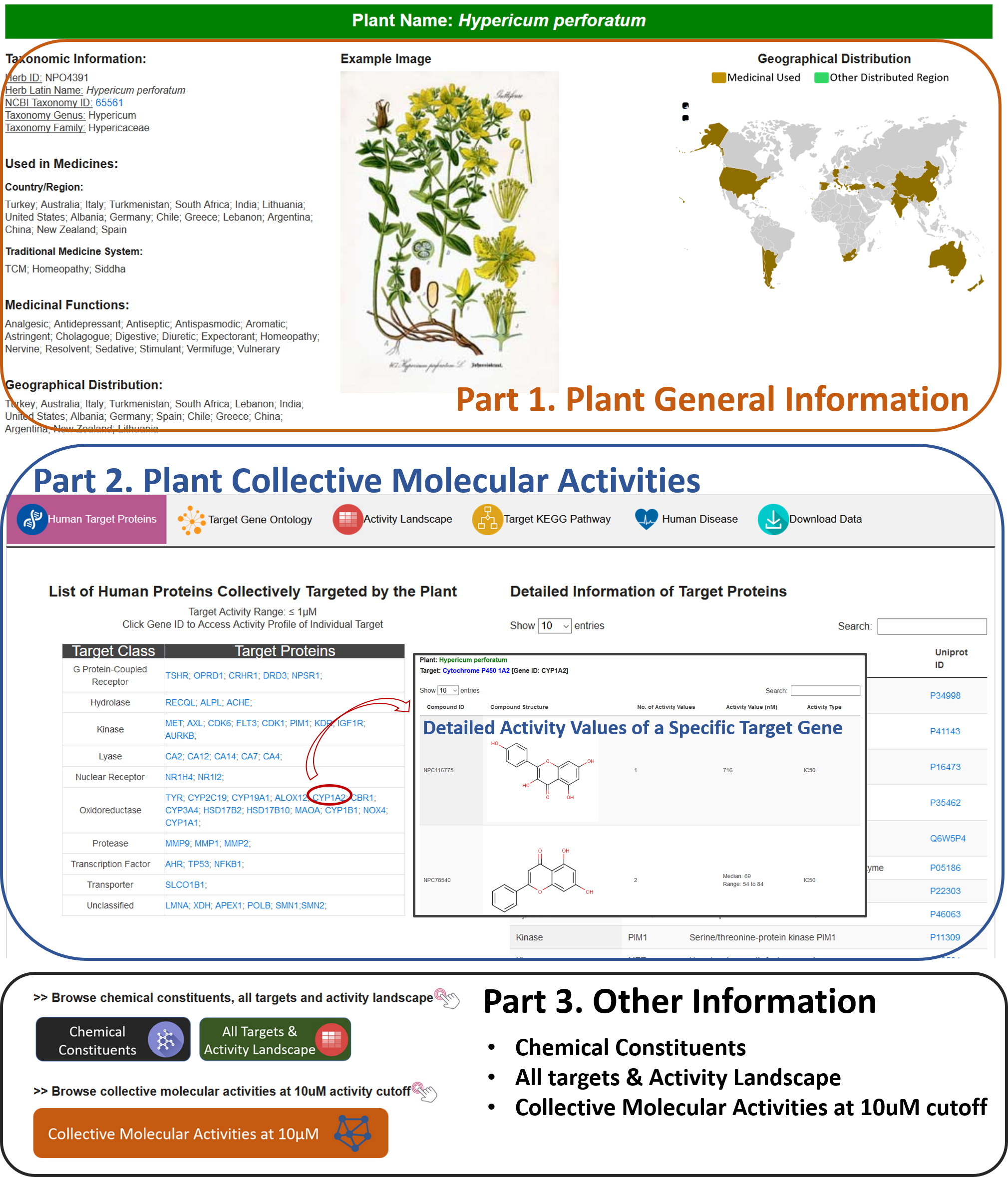
Part 1. Plant General Information Section:
In this section, the taxonomy information (species, genus, and family names) was provided, a link to NCBI Taxonomy Database was provided if available. Moreover, countries/regions or traditional medicine system where use the plant as medicinal herbs are listed if available. Geographic distribution of the plant is provided along with a highlighted world map. An example image of the plant will be displayed when available.
Part 2. Collective Molecular Activities Section:
This section includes “Human Target Genes”, “Target Gene Ontology”, “Target KEGG Pathway”, “Human Disease”, and “Download Data” subsections.
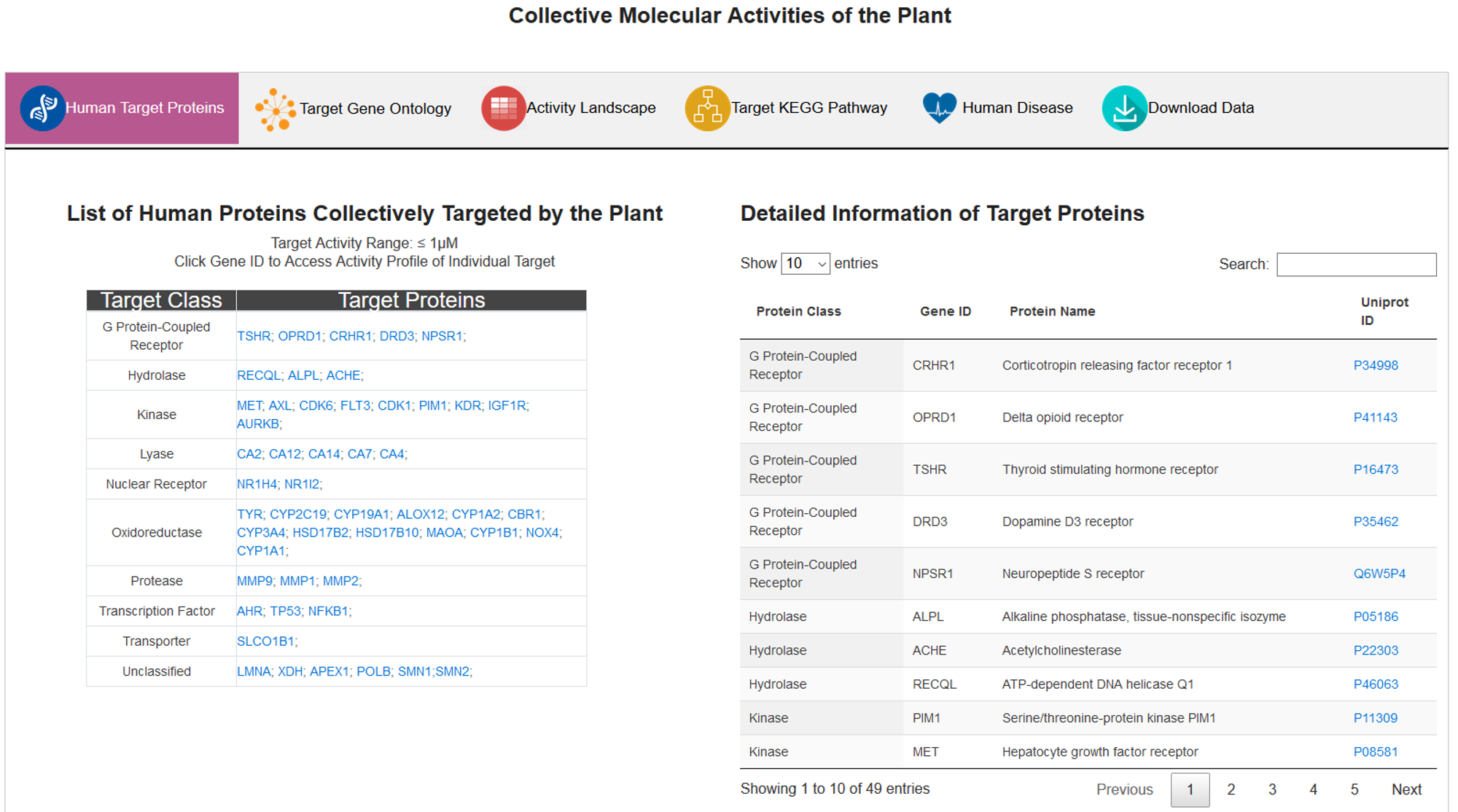
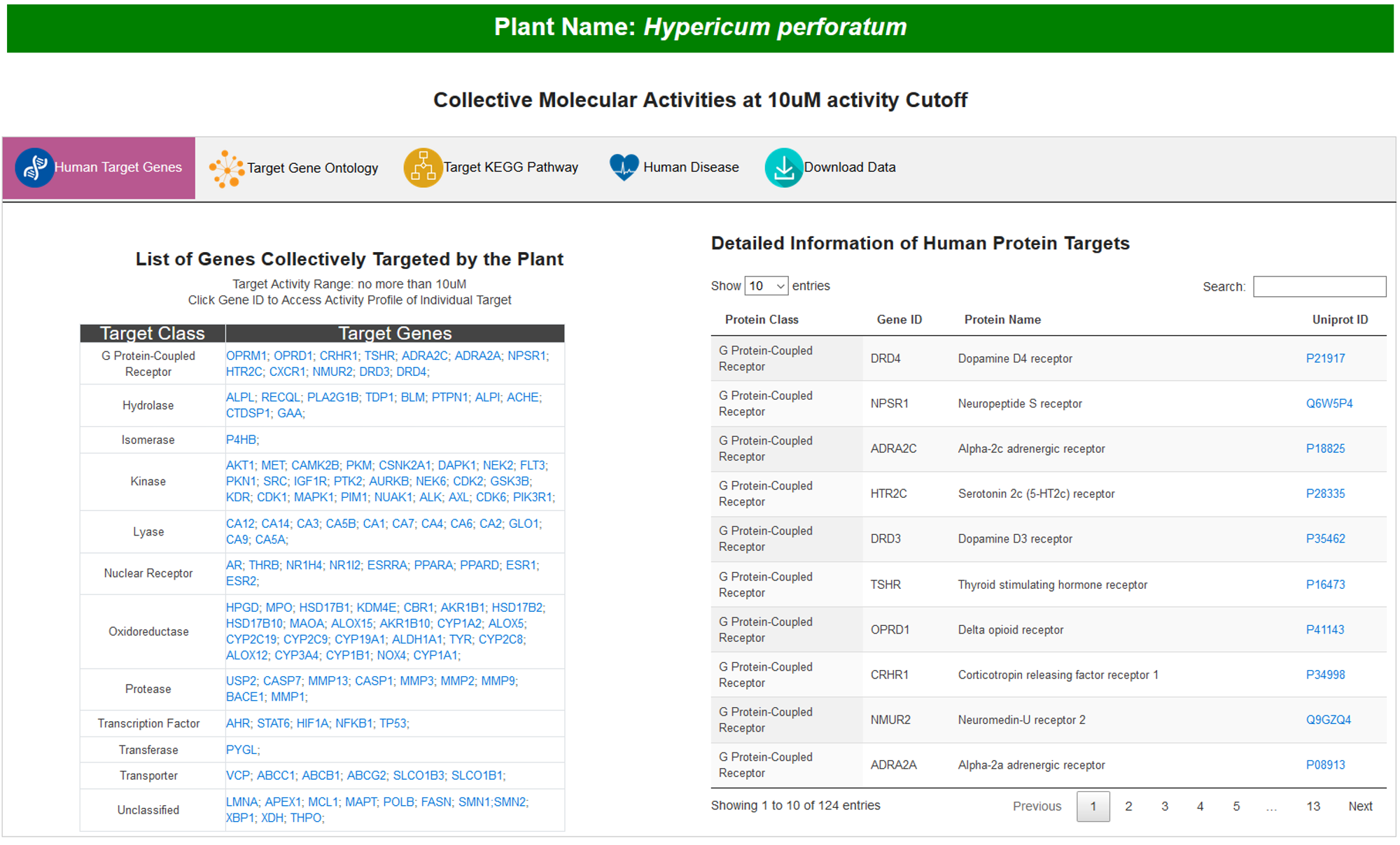
A. Human Target Genes
In this subsection, all targets are displayed in a grid graph with the highlights of target category. On the right side, the detailed information of targets is presented in an interactive table.
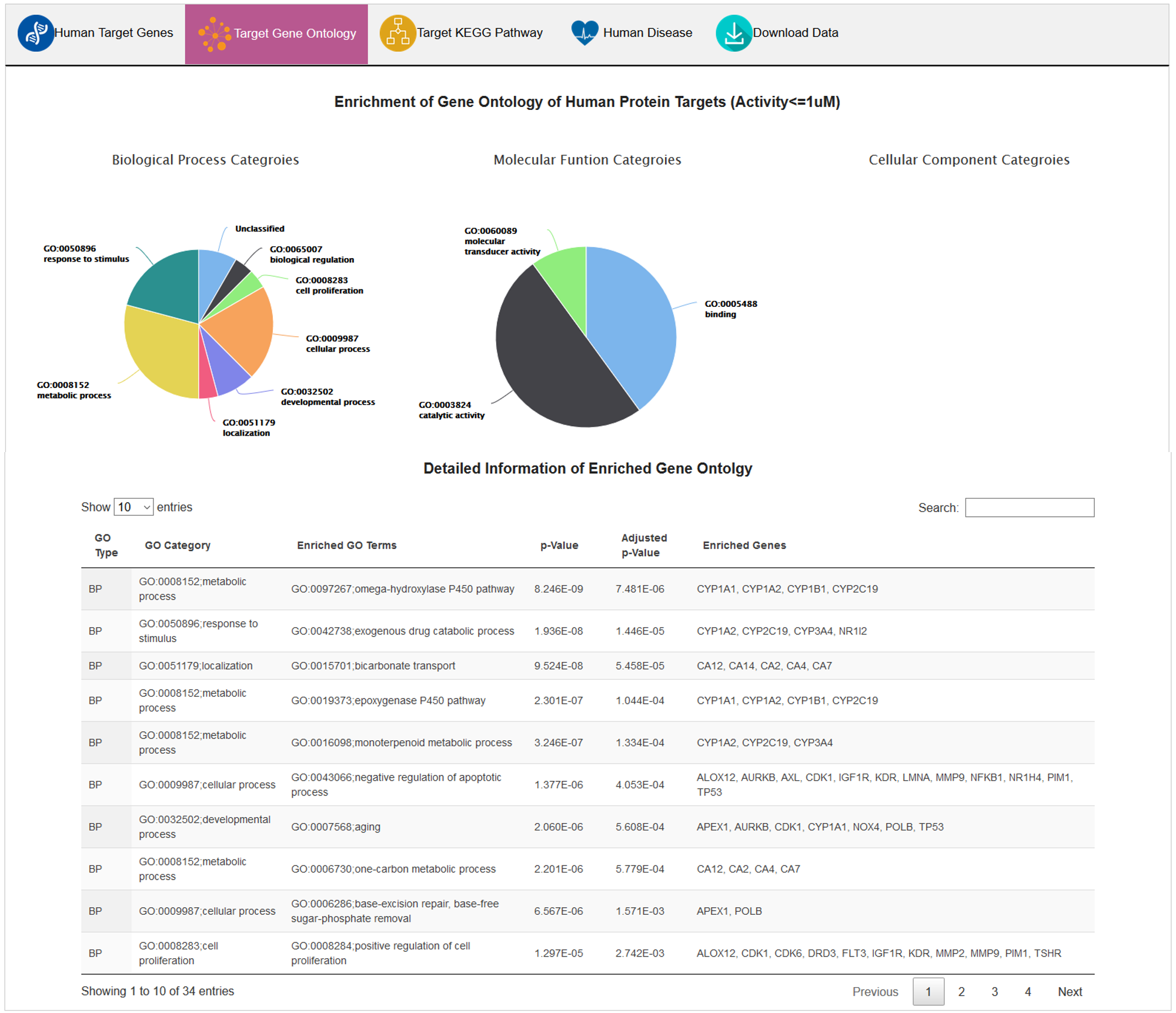
B. Target Gene Ontology
In this subsection, enriched GO terms (biological process, molecular function, and cellular component) are grouped by GO categories. Detailed data (GO term, p-value, adjusted p-value, enriched genes) was presented in an interactive table.
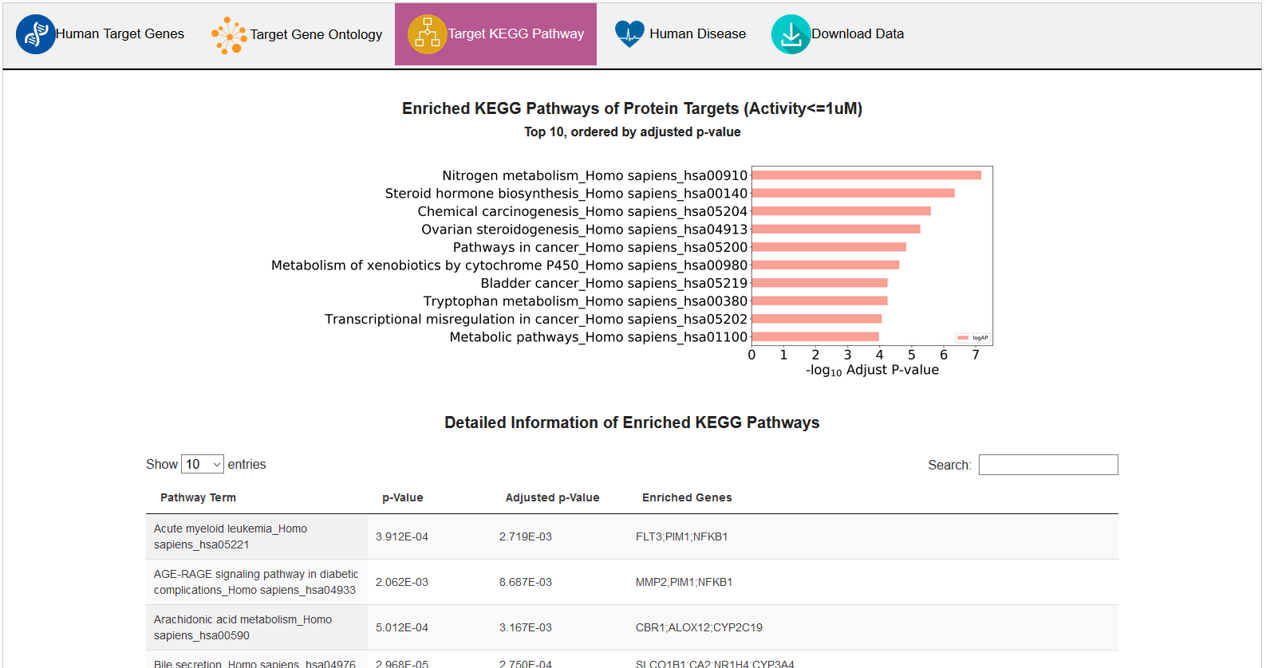
C. Target KEGG Pathway
In this subsection, top 10 enriched KEGG pathways were presented in a bar chart, detailed information was provided in the below table.
D. Human Disease
In this subsection, plant related human diseases were categorized based on ICD-10 code system of the World Health Organization.
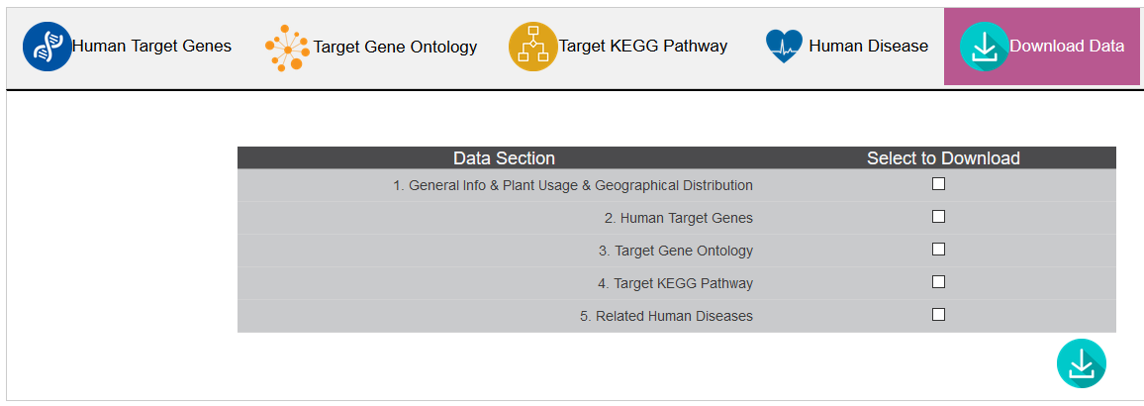
E. Download Data
Users can download all data or subsection data.
6. COLLECTIVE MOLECULAR ACTIVITIES AT 10uM ACTIVITY CUTOFF
Default activity cutoff is 1uM. Besides, collective molecular activities of plant at activity cutoff of 10uM are also provided in another webpage.
7. WEBPAGE OF PLANT’S CHEMICAL CONSTITUENTS
Chemical constituents of the plant were provided. Only those compounds which have IC50 or EC50 or Ki activity records were labelled as “Activity Available” compound part. Few carts were displayed to show the profile of several important drug-like properties.
Besides, the chemical structure images were displayed in “Compound Cards”, physico-chemical properties (calculated by using PaDEL software) were provided in another subsection.
All these data can be downloaded in the “Download Data” subsection.

8. WEBPAGE OF PLANT’S BIOLOGICAL ACTIVITY LANDSCAPE
To show the activity landscape of all ingredients of a plant, lower activity value means stronger activity and coloured with the darker red in the heatmap.
Clicking the heatmap cells can view detailed activity of certain ingredient against a specific target protein.
Moreover, users can download all the activity values in the “Download Data” subsection.
