

Who we are? Bioinformatics & Drug Design group [BIDD] is a research group based in Department of Pharmacy, Faculty of Science, National University of Singapore. Our group is active in computer-aided drug design, machine learning, pharmainformatics and cheminformatics, computational biology and bioinformatics, herbal medicine, and art and sciences.What we do? We have been developing computational methods, software tools and databases for drug discovery, target discovery, protein function prediction modeling of biological systems, network descriptor, and biomarker discovery, leading to one US patent, 14 databases, 9 web-based software tools, 3 art and science web-servers, and >250 papers published in such international journals as Nature Reviews Drug Discovery, Natural Biotechnology, PNAS, Natural Product Reports, Cancer Research, Nucleic Acids Research, and Journal of Immunology.Milestones! We pioneered inverse docking method for drug target discovery, developed the popular therapeutic target database, and are among World's first in exploring machine learning methods for protein function prediction, ADME-Tox prediction, target discovery, multi-target virtual screening.Our Alumni! BIDD alumni remain active in academic and research careers as faculty members in TongJi University, XiaMen University, ZheJiang University, Boston University, Mayo Clinic, University of Nebraska, NanJing University, SiChuan University, and National University of Singapore, as senior scientists in NIH, Harvard University, University of Georgia, National University of Singapore, A*Star IMCB and BII, and as PIs in pharmaceutical companies such as Novartis and Merk MSD.

Tenured Professor
Department of Pharmacy, Faculty of Science
National University of Singapore
Chen Yu Zong is a Tenured Professor in the Department of Pharmacy, Faculty of Science at National University of Singapore and a member of NUS Graduate School for Integrative Sciences and Engineering (NGS). He is the pioneer of 'Inverse Docking' method for drug discovery, the developer of the popular database "Therapeutic Target Database (TTD)", a researcher among World's first in exploring machine learning methods for protein function prediction, ADME-Tox prediction, target discovery, and multi-target virtual screening, the recipient of the Outstanding Scientist Award 2007 (Science Faculty, NUS). His research has led to one US patent, 14 databases, 9 web-based software tools, 3 art and science web-servers, and >250 peer-reviewed publications. Chen Yu Zong received his Ph.D. in Physics from University of Manchester, Institute of Science and Technology, U.K. in 1989. Before coming to NUS, he spent four years as Post-Doc fellow at Biophysics Group, Dept of Phys, Purdue University, Indiana, USA.
Email: Prof. Chen Yu Zong
Research Profile: Google Scholar

Postdoc Fellow
phaqc@nus.edu.sg

Postdoc Fellow
2021.08 - Now
e0021499@u.nus.edu





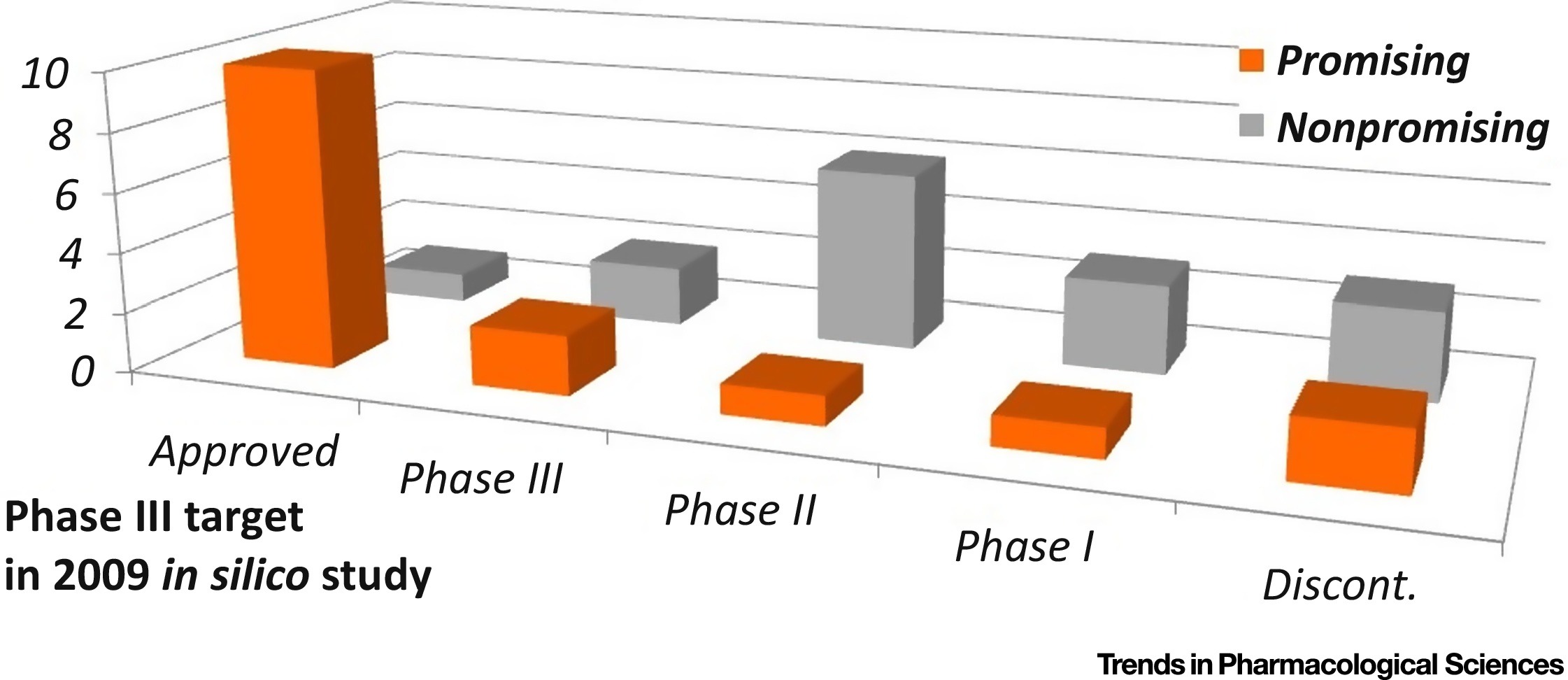 Clinical Success of Drug Targets Prospectively Predicted by In Silico Study. F. Zhu, X.X. Li, S.Y. Yang, Y. Z. Chen. Trends Pharmacol Sci. 39(3): 229-231. (2018).CellPress Clinical Success of Drug Targets Prospectively Predicted by In Silico Study. F. Zhu, X.X. Li, S.Y. Yang, Y. Z. Chen. Trends Pharmacol Sci. 39(3): 229-231. (2018).CellPress |
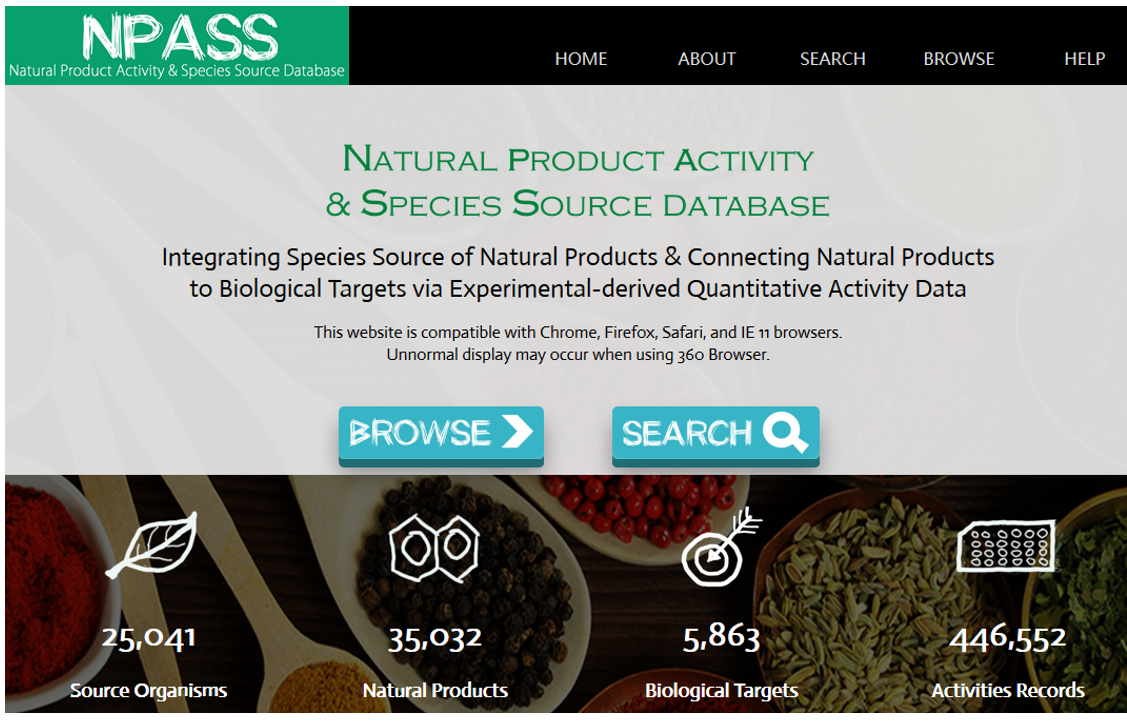 NPASS: natural product activity and species source database for natural product research, discovery and tool development. X. Zeng, P. Zhang, W.D. He, C. Qin, S.Y. Chen, L. Tao, Y. Tan, D. Gao, B.H. Wang, Z. Chen, W.P. Chen, Y.Y. Jiang, Y.Z. Chen. Nucleic Acids Res. 46(D1):D1217-D1222. (2018).PubMed NPASS: natural product activity and species source database for natural product research, discovery and tool development. X. Zeng, P. Zhang, W.D. He, C. Qin, S.Y. Chen, L. Tao, Y. Tan, D. Gao, B.H. Wang, Z. Chen, W.P. Chen, Y.Y. Jiang, Y.Z. Chen. Nucleic Acids Res. 46(D1):D1217-D1222. (2018).PubMed |
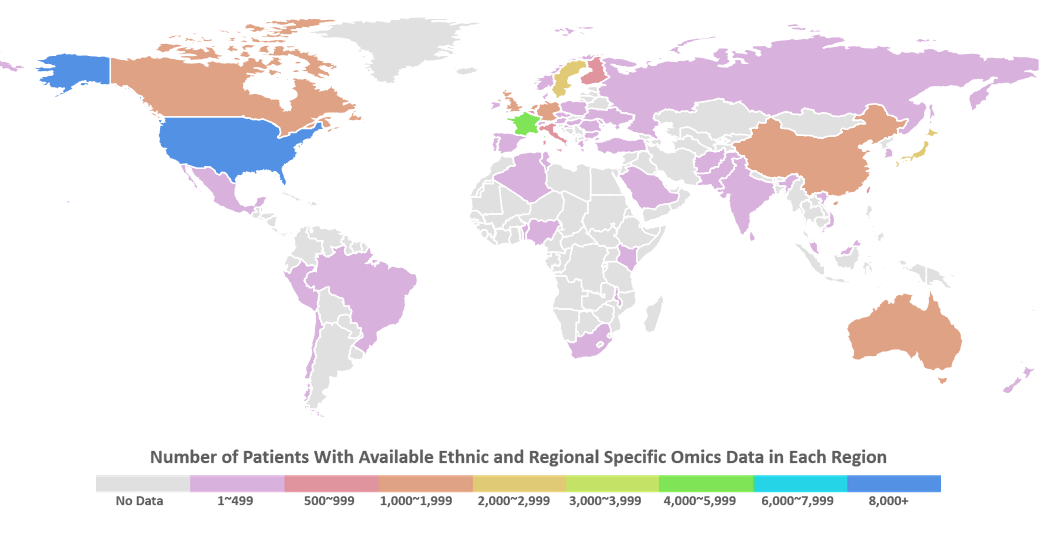 HEROD: a human ethnic and regional specific omics database. X. Zeng, L. Tao, P. Zhang, C. Qin, S. Chen, W. He, Y. Tan, H. X. Liu, S. Y. Yang, Z. Chen, Y. Y. Jiang, Y. Z. Chen. Bioinformatics. doi: 10.1093/bioinformatics/btx340 (2017).PubMed HEROD: a human ethnic and regional specific omics database. X. Zeng, L. Tao, P. Zhang, C. Qin, S. Chen, W. He, Y. Tan, H. X. Liu, S. Y. Yang, Z. Chen, Y. Y. Jiang, Y. Z. Chen. Bioinformatics. doi: 10.1093/bioinformatics/btx340 (2017).PubMed |
 A protein network descriptor server and its use in studying protein, disease, metabolic and drug targeted networks. P. Zhang, L. Tao, X. Zeng, C. Qin, S.Y. Chen, F. Zhu, Z.R. Li, Y.Y. Jiang, W.P. Chen, Y.Z. Chen.Brief Bioinform. pii: bbw071 (2016).PubMed A protein network descriptor server and its use in studying protein, disease, metabolic and drug targeted networks. P. Zhang, L. Tao, X. Zeng, C. Qin, S.Y. Chen, F. Zhu, Z.R. Li, Y.Y. Jiang, W.P. Chen, Y.Z. Chen.Brief Bioinform. pii: bbw071 (2016).PubMed |
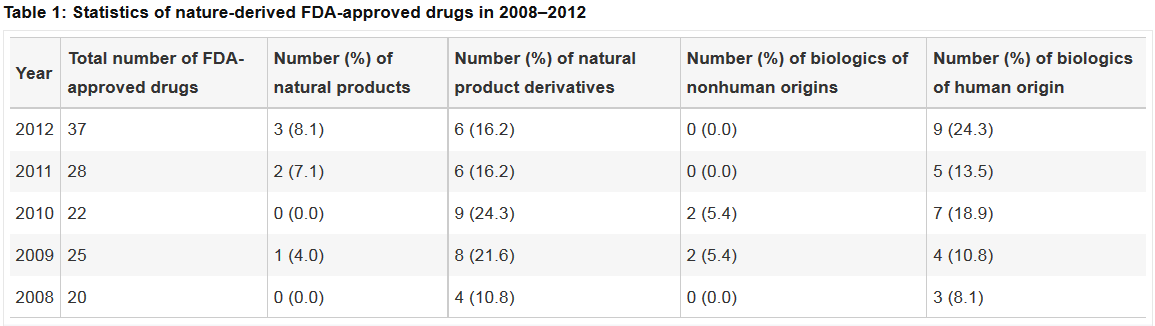 Nature's contribution to today's pharmacopeia. L. Tao, F. Zhu, C. Qin, C. Zhang, F. Xu, C.Y. Tan, Y.Y. Jiang, Y.Z. Chen. Nat Biotechnol. 32(10):979-80 (2014).PubMed Nature's contribution to today's pharmacopeia. L. Tao, F. Zhu, C. Qin, C. Zhang, F. Xu, C.Y. Tan, Y.Y. Jiang, Y.Z. Chen. Nat Biotechnol. 32(10):979-80 (2014).PubMed |
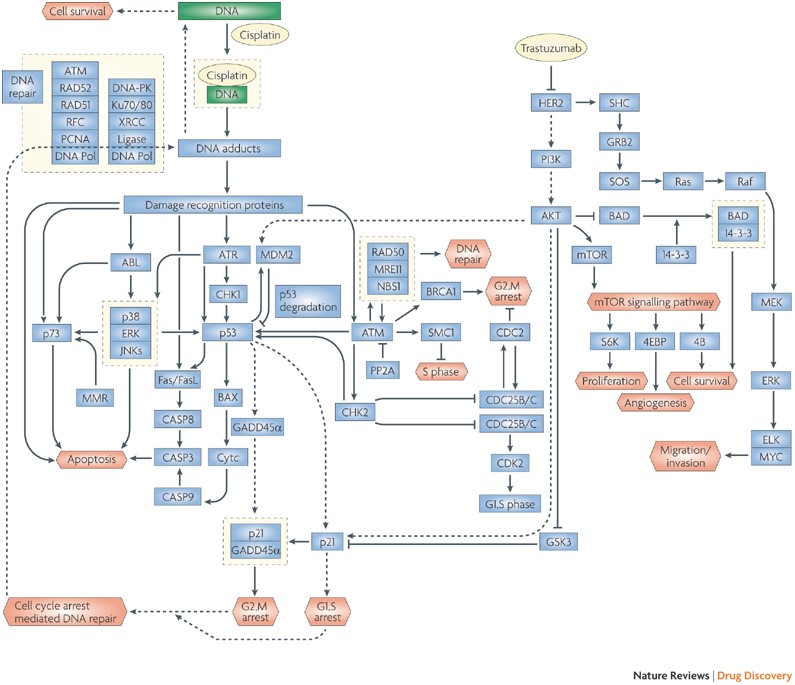 Mechanisms of drug combinations from interaction and network perspectives J. Jia, F. Zhu, X.H. Ma, Z.W. Cao, Y.X. Li and Y.Z. Chen. Nat. Rev. Drug Discov. 8(2):111-28(2009).PubMed Mechanisms of drug combinations from interaction and network perspectives J. Jia, F. Zhu, X.H. Ma, Z.W. Cao, Y.X. Li and Y.Z. Chen. Nat. Rev. Drug Discov. 8(2):111-28(2009).PubMed |







