Plant Name: Persicaria tinctoria
Taxonomic Information:
Plant ID: NPO9069
Plant Latin Name: Persicaria tinctoria
Taxonomy Genus: Persicaria
Taxonomy Family: Polygonaceae
Plant External Links:
NCBI TaxonomyDB:
96455
Plant-of-the-World-Online:
60455325-2
Used in Medicines:
Country/Region:
South KoreaTraditional Medicine System:
Geographical Distribution:
Ukraine; South Korea; China; South Africa
Collective Molecular Activities of the Plant
List of Human Proteins Collectively Targeted by the Plant
Click Gene ID to Access Activity Profile of Individual Target
| ADORA3; ADORA2A; ADORA1; ADRA2B; | |
| TSHR; NPSR1; | |
| CDA; ADK; ALOX12; HSD17B2; HSD17B10; NOX4; ALOX15; POLB; | |
| MET; AXL; FLT3; CDK1; PIM1; KDR; IGF1R; AURKB; | |
| CA2; CA12; CA7; CA4; | |
| PPARA; PPARD; | |
| ALOX5; TYR; PTGS2; | |
| MMP9; MMP1; MMP2; | |
| AHR; NFKB1; | |
| LMNA; |
List of Human Cytochrome P450 Enzymes Affected by the Plant
Click Gene ID to Access Activity Profile of Individual Enzyme
Detailed Information of Target Proteins
| Protein Class | Gene ID | Protein Name | Uniprot ID | Target ChEMBL ID |
|---|---|---|---|---|
| Cytochrome P450 family 1 | CYP1A2 | Cytochrome P450 1A2 | P05177 | CHEMBL3356 |
| Cytochrome P450 family 1 | CYP1B1 | Cytochrome P450 1B1 | Q16678 | CHEMBL4878 |
| Cytochrome P450 family 1 | CYP1A1 | Cytochrome P450 1A1 | P04798 | CHEMBL2231 |
| Enzyme_unclassified | ALOX12 | Arachidonate 12-lipoxygenase | P18054 | CHEMBL3687 |
| Enzyme_unclassified | HSD17B2 | Estradiol 17-beta-dehydrogenase 2 | P37059 | CHEMBL2789 |
| Enzyme_unclassified | HSD17B10 | Endoplasmic reticulum-associated amyloid beta-peptide-binding protein | Q99714 | CHEMBL4159 |
| Enzyme_unclassified | NOX4 | NADPH oxidase 4 | Q9NPH5 | CHEMBL1250375 |
| Enzyme_unclassified | ALOX15 | Arachidonate 15-lipoxygenase | P16050 | CHEMBL2903 |
| Enzyme_unclassified | POLB | DNA polymerase beta | P06746 | CHEMBL2392 |
| Enzyme_unclassified | CDA | Cytidine deaminase | P32320 | CHEMBL4502 |
Showing 1 to 10 of 40 entries
Enrichment of Gene Ontology of Target Proteins (Activity≤1μM)
Detailed Information of Enriched Gene Ontolgy
| GO Type | GO Category | Enriched GO Terms | p-Value | Adjusted p-Value | Enriched Genes |
|---|---|---|---|---|---|
| BP | GO:0009987; cellular process | GO:0043066; negative regulation of apoptotic process | 8.150E-10 | 1.471E-06 | ADORA1, ADORA2A, ALOX12, AURKB, AXL, CDK1, IGF1R, KDR, LMNA, MMP9, NFKB1, PIM1, PPARD, PTGS2 |
| BP | GO:0008152; metabolic process | GO:0019372; lipoxygenase pathway | 2.818E-08 | 1.979E-05 | ALOX12, ALOX15, ALOX5, PTGS2 |
| BP | GO:0050896; response to stimulus | GO:0001666; response to hypoxia | 4.977E-08 | 2.929E-05 | ADORA1, CYP1A1, LMNA, MMP2, NOX4, PPARA, PPARD, PTGS2 |
| BP | GO:0008152; metabolic process | GO:0055114; oxidation-reduction process | 1.585E-07 | 7.345E-05 | ALOX12, ALOX15, ALOX5, CYP1A1, CYP1A2, CYP1B1, HSD17B10, HSD17B2, NOX4, PPARD, PTGS2, TYR |
| BP | GO:0009987; cellular process | GO:0001973; adenosine receptor signaling pathway | 6.851E-07 | 2.162E-04 | ADORA1, ADORA2A, ADORA3 |
| BP | GO:0008152; metabolic process | GO:0006730; one-carbon metabolic process | 8.851E-07 | 2.667E-04 | CA12, CA2, CA4, CA7 |
| BP | GO:0008152; metabolic process | GO:0097267; omega-hydroxylase P450 pathway | 9.773E-07 | 2.876E-04 | CYP1A1, CYP1A2, CYP1B1 |
| BP | GO:0022414; reproductive process | GO:0007566; embryo implantation | 1.106E-06 | 3.168E-04 | MMP2, MMP9, PPARD, PTGS2 |
| BP | GO:0051179; localization | GO:0015701; bicarbonate transport | 2.016E-06 | 5.225E-04 | CA12, CA2, CA4, CA7 |
| BP | GO:0023052; signaling | GO:0032230; positive regulation of synaptic transmission, GABAergic | 2.319E-06 | 5.805E-04 | ADORA2A, CA2, CA7 |
Showing 1 to 10 of 46 entries
Enriched KEGG Pathways of Target Proteins (Activity≤1μM)
Top 10 KEGG Pathways (ordered by adjusted p-value)
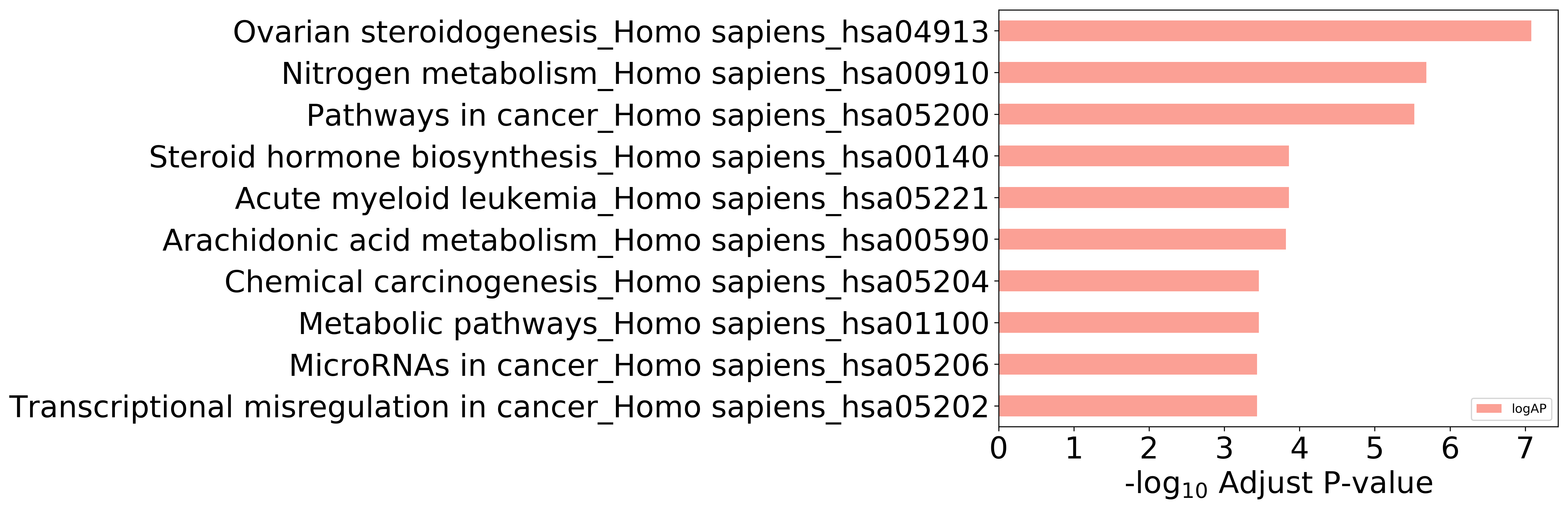
Detailed Information of Enriched KEGG Pathways
| Pathway Category Top Level | Pathway Category Second Level | Pathway ID | Pathway Name | p-Value | Adjusted p-Value | Enriched Genes |
|---|---|---|---|---|---|---|
| 09100 Metabolism | 09102 Energy metabolism | hsa00910 | Nitrogen metabolism | 3.203E-08 | 2.066E-06 | CA12, CA2, CA4, CA7 |
| 09100 Metabolism | 09103 Lipid metabolism | hsa00590 | Arachidonic acid metabolism | 7.036E-06 | 1.513E-04 | ALOX5, ALOX15, ALOX12, PTGS2 |
| 09100 Metabolism | 09103 Lipid metabolism | hsa00140 | Steroid hormone biosynthesis | 5.382E-06 | 1.389E-04 | CYP1A2, HSD17B2, CYP1A1, CYP1B1 |
| 09100 Metabolism | 09105 Amino acid metabolism | hsa00380 | Tryptophan metabolism | 6.956E-05 | 6.445E-04 | CYP1A2, CYP1A1, CYP1B1 |
| 09100 Metabolism | 09111 Xenobiotics biodegradation and metabolism | hsa00980 | Metabolism of xenobiotics by cytochrome P450 | 4.183E-04 | 2.840E-03 | CYP1A2, CYP1A1, CYP1B1 |
| 09100 Metabolism | 09103 Lipid metabolism | hsa00591 | Linoleic acid metabolism | 1.530E-03 | 7.896E-03 | CYP1A2, ALOX15 |
| 09130 Environmental Information Processing | 09132 Signal transduction | hsa04024 | cAMP signaling pathway | 4.598E-05 | 5.392E-04 | ADORA2A, ADORA1, PPARA, NFKB1, TSHR |
| 09130 Environmental Information Processing | 09133 Signaling molecules and interaction | hsa04080 | Neuroactive ligand-receptor interaction | 2.175E-04 | 1.651E-03 | ADORA2A, ADORA3, ADORA1, ADRA2B, TSHR |
| 09130 Environmental Information Processing | 09132 Signal transduction | hsa04015 | Rap1 signaling pathway | 8.172E-04 | 5.271E-03 | ADORA2A, KDR, MET, IGF1R |
| 09130 Environmental Information Processing | 09132 Signal transduction | hsa04014 | Ras signaling pathway | 1.072E-03 | 6.048E-03 | KDR, MET, NFKB1, IGF1R |
Showing 1 to 10 of 26 entries
Human Diseases Associated with the Target Proteins of the Plant
Detailed Information of Human Diseases
| ICD10 Disease Category | Disease Name | ICD10 Code | Associated Disease Targets (Association Ref: TTD database) |
|---|---|---|---|
| A00-B99: Certain infectious and parasitic diseases | Lymphatic filariasis | B74 | ALOX5; |
| A00-B99: Certain infectious and parasitic diseases | Malaria | B54 | ADORA3; |
| A00-B99: Certain infectious and parasitic diseases | Human immunodeficiency virus infection | B20-B26 | FLT3; |
| A00-B99: Certain infectious and parasitic diseases | Sepsis | A40, A41 | ADORA1; |
| A00-B99: Certain infectious and parasitic diseases | Fungal infections | B35-B49 | ALOX5; |
| C00-D49: Neoplasms | Head and neck cancer | C07-C14, C32-C33 | KDR; |
| C00-D49: Neoplasms | Thyroid cancer diagnosis | C73 | TSHR; |
| C00-D49: Neoplasms | Non-small cell lung cancer | C33-C34 | MET; IGF1R; KDR; AXL; PTGS2; |
| C00-D49: Neoplasms | Inflammatory disease | C11, C44, K75.9, M00-M25 | PTGS2; ALOX5; |
| C00-D49: Neoplasms | Metastatic breast cancer | C50 | KDR; |
Showing 1 to 10 of 153 entries


