Plant Name: Nama hispidum
Collective Molecular Activities of the Plant
List of Human Proteins Collectively Targeted by the Plant
Click Gene ID to Access Activity Profile of Individual Target
| GPR35; | |
| NPSR1; | |
| RECQL; TDP1; TERT; GLO1; PGD; ALOX12; AKR1B1; HSD17B2; HSD17B10; HPGD; NOX4; APEX1; POLB; | |
| ACHE; | |
| BCL2; | |
| TOP2A; | |
| TEK; INSR; MET; AXL; KDR; FLT3; CDK1; EGFR; PIM1; NUAK1; SRC; IGF1R; AURKB; CSNK2A1; | |
| CA2; CA12; CA7; CA4; | |
| TYR; | |
| KDM4E; | |
| F10; F2; | |
| AHR; | |
| FUT7; | |
| DNMT1; | |
| ABCB1; | |
| SLC22A6; | |
| LMNA; SMAD3; HSPA1A; | |
| L3MBTL1; |
List of Human Cytochrome P450 Enzymes Affected by the Plant
Click Gene ID to Access Activity Profile of Individual Enzyme
Detailed Information of Target Proteins
| Protein Class | Gene ID | Protein Name | Uniprot ID | Target ChEMBL ID |
|---|---|---|---|---|
| ATP-binding cassette | ABCB1 | P-glycoprotein 1 | P08183 | CHEMBL4302 |
| Cytochrome P450 family 1 | CYP1B1 | Cytochrome P450 1B1 | Q16678 | CHEMBL4878 |
| Cytochrome P450 family 1 | CYP1A1 | Cytochrome P450 1A1 | P04798 | CHEMBL2231 |
| Cytochrome P450 family 1 | CYP1A2 | Cytochrome P450 1A2 | P05177 | CHEMBL3356 |
| DNA methyltransferase | DNMT1 | DNA (cytosine-5)-methyltransferase 1 | P26358 | CHEMBL1993 |
| Enzyme_unclassified | RECQL | ATP-dependent DNA helicase Q1 | P46063 | CHEMBL1293236 |
| Enzyme_unclassified | TDP1 | Tyrosyl-DNA phosphodiesterase 1 | Q9NUW8 | CHEMBL1075138 |
| Enzyme_unclassified | TERT | Telomerase reverse transcriptase | O14746 | CHEMBL2916 |
| Enzyme_unclassified | GLO1 | Glyoxalase I | Q04760 | CHEMBL2424 |
| Enzyme_unclassified | PGD | 6-phosphogluconate dehydrogenase | P52209 | CHEMBL3404 |
Showing 1 to 10 of 52 entries
Enrichment of Gene Ontology of Target Proteins (Activity≤1μM)
Detailed Information of Enriched Gene Ontolgy
| GO Type | GO Category | Enriched GO Terms | p-Value | Adjusted p-Value | Enriched Genes |
|---|---|---|---|---|---|
| BP | GO:0009987; cellular process | GO:0051897; positive regulation of protein kinase B signaling | 6.858E-09 | 2.588E-06 | AXL, EGFR, F10, INSR, MET, NOX4, SRC, TEK |
| BP | GO:0008152; metabolic process | GO:0038083; peptidyl-tyrosine autophosphorylation | 1.635E-07 | 3.632E-05 | EGFR, IGF1R, INSR, KDR, SRC |
| BP | GO:0050896; response to stimulus | GO:0009611; response to wounding | 4.911E-07 | 9.141E-05 | ACHE, BCL2, CDK1, CYP1A1, EGFR, F2, SMAD3 |
| BP | GO:0008152; metabolic process | GO:0055114; oxidation-reduction process | 5.108E-07 | 9.426E-05 | AKR1B1, ALOX12, APEX1, CYP1A1, CYP1A2, CYP1B1, HPGD, HSD17B10, HSD17B2, KDM4E, NOX4, PGD, TYR |
| BP | GO:0009987; cellular process | GO:0070301; cellular response to hydrogen peroxide | 6.162E-07 | 1.082E-04 | AKR1B1, APEX1, AXL, CDK1, CYP1B1 |
| BP | Unclassified; | GO:2000021; regulation of ion homeostasis | 1.346E-06 | 2.109E-04 | BCL2, CA2, CA7, F2, KDR, NPSR1, SRC |
| BP | GO:0032502; developmental process | GO:0031100; animal organ regeneration | 1.856E-06 | 2.750E-04 | AXL, CDK1, CSNK2A1, EGFR, FLT3 |
| BP | GO:0051179; localization | GO:2001225; regulation of chloride transport | 2.179E-06 | 3.163E-04 | ABCB1, CA2, CA7 |
| BP | GO:0008152; metabolic process | GO:0097267; omega-hydroxylase P450 pathway | 2.179E-06 | 3.163E-04 | CYP1A1, CYP1A2, CYP1B1 |
| BP | GO:0040011; locomotion | GO:0014911; positive regulation of smooth muscle cell migration | 2.581E-06 | 3.649E-04 | BCL2, NOX4, SRC, TERT |
Showing 1 to 10 of 54 entries
Enriched KEGG Pathways of Target Proteins (Activity≤1μM)
Top 10 KEGG Pathways (ordered by adjusted p-value)
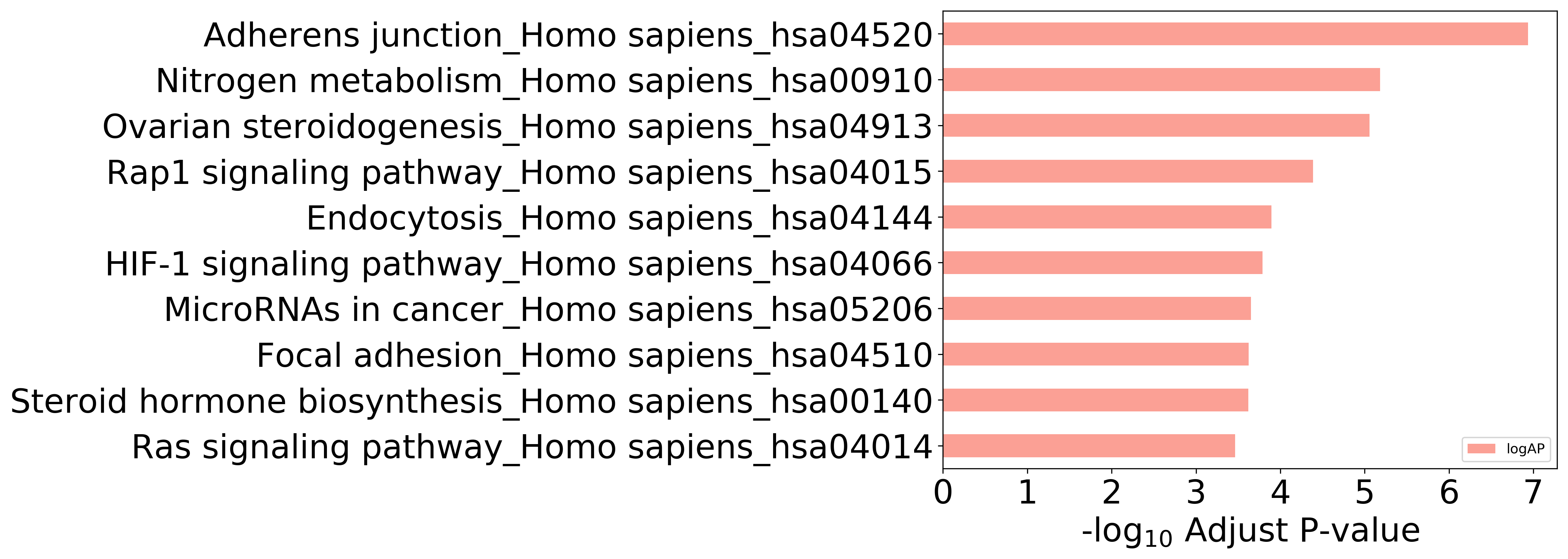
Detailed Information of Enriched KEGG Pathways
| Pathway Category Top Level | Pathway Category Second Level | Pathway ID | Pathway Name | p-Value | Adjusted p-Value | Enriched Genes |
|---|---|---|---|---|---|---|
| 09100 Metabolism | 09102 Energy metabolism | hsa00910 | Nitrogen metabolism | 9.429E-08 | 6.553E-06 | CA12, CA2, CA4, CA7 |
| 09100 Metabolism | 09103 Lipid metabolism | hsa00140 | Steroid hormone biosynthesis | 1.554E-05 | 2.400E-04 | CYP1A2, HSD17B2, CYP1A1, CYP1B1 |
| 09100 Metabolism | 09105 Amino acid metabolism | hsa00380 | Tryptophan metabolism | 1.530E-04 | 1.773E-03 | CYP1A2, CYP1A1, CYP1B1 |
| 09100 Metabolism | 09111 Xenobiotics biodegradation and metabolism | hsa00980 | Metabolism of xenobiotics by cytochrome P450 | 9.068E-04 | 6.302E-03 | CYP1A2, CYP1A1, CYP1B1 |
| 09130 Environmental Information Processing | 09132 Signal transduction | hsa04015 | Rap1 signaling pathway | 1.178E-06 | 4.093E-05 | SRC, INSR, KDR, TEK, MET, EGFR, IGF1R |
| 09130 Environmental Information Processing | 09132 Signal transduction | hsa04066 | HIF-1 signaling pathway | 7.045E-06 | 1.632E-04 | INSR, BCL2, TEK, EGFR, IGF1R |
| 09130 Environmental Information Processing | 09132 Signal transduction | hsa04151 | PI3K-Akt signaling pathway | 2.726E-05 | 3.445E-04 | INSR, KDR, BCL2, TEK, MET, EGFR, IGF1R |
| 09130 Environmental Information Processing | 09132 Signal transduction | hsa04014 | Ras signaling pathway | 2.634E-05 | 3.445E-04 | INSR, KDR, TEK, MET, EGFR, IGF1R |
| 09130 Environmental Information Processing | 09132 Signal transduction | hsa04068 | FoxO signaling pathway | 3.952E-04 | 3.923E-03 | SMAD3, INSR, EGFR, IGF1R |
| 09140 Cellular Processes | 09144 Cellular community - eukaryotes | hsa04520 | Adherens junction | 8.315E-10 | 1.156E-07 | SMAD3, CSNK2A1, SRC, INSR, MET, EGFR, IGF1R |
Showing 1 to 10 of 27 entries
Human Diseases Associated with the Target Proteins of the Plant
Detailed Information of Human Diseases
| ICD10 Disease Category | Disease Name | ICD10 Code | Associated Disease Targets (Association Ref: TTD database) |
|---|---|---|---|
| A00-B99: Certain infectious and parasitic diseases | Pediculus humanus capitis | B85.0 | ACHE; |
| A00-B99: Certain infectious and parasitic diseases | Cryptosporidium infection | A07.2 | EGFR; |
| A00-B99: Certain infectious and parasitic diseases | Human immunodeficiency virus infection | B20-B26 | FLT3; |
| A00-B99: Certain infectious and parasitic diseases | Helminth infection | A00-B99 | ACHE; |
| A00-B99: Certain infectious and parasitic diseases | Bacterial infections | A00-B99 | ABCB1; |
| C00-D49: Neoplasms | Non-small-cell lung cancer | NA | KDR; |
| C00-D49: Neoplasms | Non-small cell lung cancer | C33-C34 | MET; IGF1R; KDR; AXL; DNMT1; EGFR; ABCB1; |
| C00-D49: Neoplasms | Refractory breast cancer | C50 | EGFR; |
| C00-D49: Neoplasms | Renal cell carcinoma | NA | KDR; |
| C00-D49: Neoplasms | Chronic lymphocytic leukaemia | C91 | EGFR; BCL2; |
Showing 1 to 10 of 173 entries


