Plant Name: Alnus firma
Taxonomic Information:
Plant ID: NPO4971
Plant Latin Name: Alnus firma
Taxonomy Genus: Alnus
Taxonomy Family: Betulaceae
Plant External Links:
NCBI TaxonomyDB:
109059
Plant-of-the-World-Online:
n.a.
Collective Molecular Activities of the Plant
List of Human Proteins Collectively Targeted by the Plant
Click Gene ID to Access Activity Profile of Individual Target
| GPBAR1; | |
| PTPN1; PTPN2; CDC25B; | |
| PLA2G1B; AKR1B10; POLB; | |
| NR1H4; |
List of Human Cytochrome P450 Enzymes Affected by the Plant
Click Gene ID to Access Activity Profile of Individual Enzyme
| Cytochrome P450 Enzymes: |
|---|
Detailed Information of Target Proteins
| Protein Class | Gene ID | Protein Name | Uniprot ID | Target ChEMBL ID |
|---|---|---|---|---|
| Enzyme_unclassified | PLA2G1B | Phospholipase A2 group 1B | P04054 | CHEMBL4426 |
| Enzyme_unclassified | AKR1B10 | Aldo-keto reductase family 1 member B10 | O60218 | CHEMBL5983 |
| Enzyme_unclassified | POLB | DNA polymerase beta | P06746 | CHEMBL2392 |
| Nuclear hormone receptor subfamily 1 group H | NR1H4 | Bile acid receptor FXR | Q96RI1 | CHEMBL2047 |
| Protein Phosphatase | PTPN1 | Protein-tyrosine phosphatase 1B | P18031 | CHEMBL335 |
| Protein Phosphatase | PTPN2 | T-cell protein-tyrosine phosphatase | P17706 | CHEMBL3807 |
| Protein Phosphatase | CDC25B | Dual specificity phosphatase Cdc25B | P30305 | CHEMBL4804 |
| Small molecule receptor (family A GPCR) | GPBAR1 | G-protein coupled bile acid receptor 1 | Q8TDU6 | CHEMBL5409 |
Enrichment of Gene Ontology of Target Proteins (Activity≤10μM)
Detailed Information of Enriched Gene Ontolgy
| GO Type | GO Category | Enriched GO Terms | p-Value | Adjusted p-Value | Enriched Genes |
|---|---|---|---|---|---|
| BP | GO:0009987; cellular process | GO:0038183; bile acid signaling pathway | 4.502E-07 | 4.901E-03 | GPBAR1, NR1H4 |
| BP | GO:0050896; response to stimulus | GO:0046626; regulation of insulin receptor signaling pathway | 8.514E-07 | 4.901E-03 | NR1H4, PTPN1, PTPN2 |
| BP | GO:0009987; cellular process | GO:1902202; regulation of hepatocyte growth factor receptor signaling pathway | 9.002E-07 | 4.901E-03 | PTPN1, PTPN2 |
| MF | GO:0060089; molecular transducer activity | GO:0038181; bile acid receptor activity | 4.502E-07 | 4.901E-03 | GPBAR1, NR1H4 |
| BP | GO:0008152; metabolic process | GO:0016311; dephosphorylation | 3.058E-06 | 1.110E-02 | CDC25B, POLB, PTPN1, PTPN2 |
| MF | GO:0005488; binding | GO:0032052; bile acid binding | 5.396E-06 | 1.678E-02 | NR1H4, PLA2G1B |
| BP | GO:0009987; cellular process | GO:1903897; regulation of PERK-mediated unfolded protein response | 6.743E-06 | 1.710E-02 | PTPN1, PTPN2 |
| BP | GO:0008152; metabolic process | GO:0035335; peptidyl-tyrosine dephosphorylation | 7.178E-06 | 1.710E-02 | CDC25B, PTPN1, PTPN2 |
| MF | GO:0003824; catalytic activity | GO:0004725; protein tyrosine phosphatase activity | 7.854E-06 | 1.710E-02 | CDC25B, PTPN1, PTPN2 |
| BP | GO:0009987; cellular process | GO:1900103; positive regulation of endoplasmic reticulum unfolded protein response | 1.168E-05 | 2.312E-02 | PTPN1, PTPN2 |
| BP | GO:0009987; cellular process | GO:0010804; negative regulation of tumor necrosis factor-mediated signaling pathway | 2.841E-05 | 5.156E-02 | NR1H4, PTPN2 |
| BP | GO:0009987; cellular process | GO:0001959; regulation of cytokine-mediated signaling pathway | 3.443E-05 | 5.747E-02 | NR1H4, PTPN1, PTPN2 |
| MF | GO:0005488; binding | GO:0030971; receptor tyrosine kinase binding | 2.286E-04 | 1.478E-01 | PTPN1, PTPN2 |
| CC | GO:0044464; cell part | GO:0097443; sorting endosome | 2.483E-03 | 3.888E-01 | PTPN1 |
| CC | GO:0043226; organelle | GO:0005876; spindle microtubule | 2.174E-02 | 1.000E+00 | POLB |
| CC | GO:0043226; organelle | GO:0005654; nucleoplasm | 2.458E-02 | 1.000E+00 | CDC25B, NR1H4, POLB, PTPN2 |
| CC | GO:0044464; cell part | GO:0005829; cytosol | 2.751E-02 | 1.000E+00 | AKR1B10, CDC25B, PLA2G1B, PTPN1, PTPN2 |
Enriched KEGG Pathways of Target Proteins (Activity≤10μM)
Top 10 KEGG Pathways (ordered by adjusted p-value)
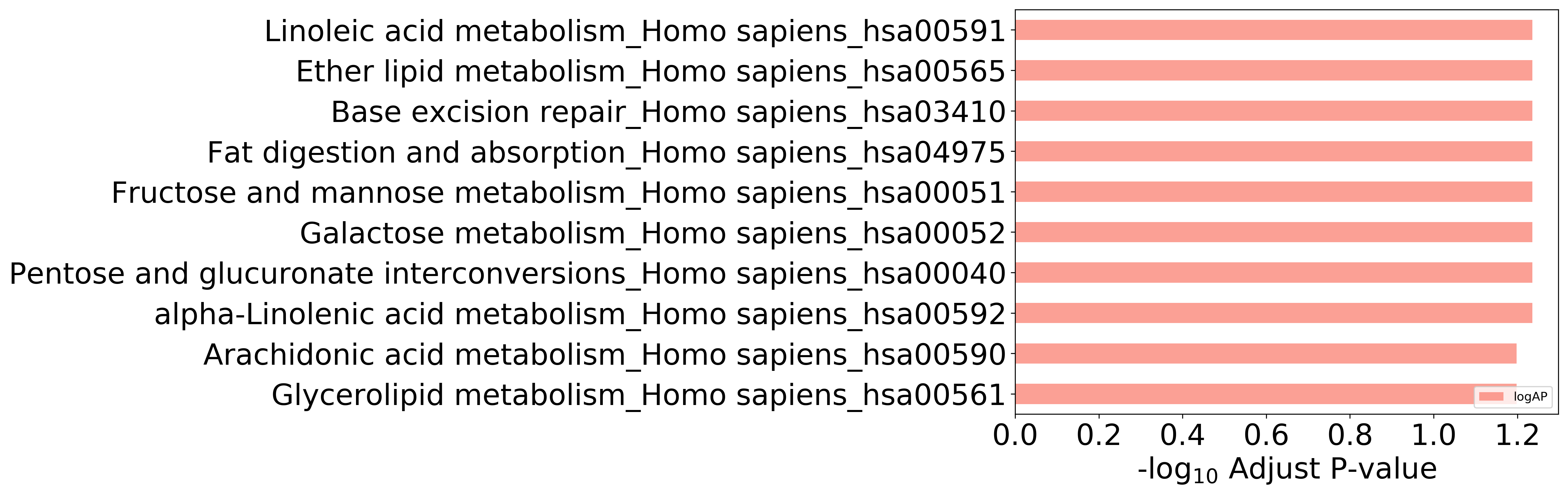
Detailed Information of Enriched KEGG Pathways
| Pathway Category Top Level | Pathway Category Second Level | Pathway ID | Pathway Name | p-Value | Adjusted p-Value | Enriched Genes |
|---|---|---|---|---|---|---|
| 09100 Metabolism | 09103 Lipid metabolism | hsa00592 | alpha-Linolenic acid metabolism | 9.958E-03 | 5.805E-02 | PLA2G1B |
| 09100 Metabolism | 09103 Lipid metabolism | hsa00591 | Linoleic acid metabolism | 1.154E-02 | 5.805E-02 | PLA2G1B |
| 09100 Metabolism | 09101 Carbohydrate metabolism | hsa00040 | Pentose and glucuronate interconversions | 1.431E-02 | 5.805E-02 | AKR1B10 |
| 09100 Metabolism | 09101 Carbohydrate metabolism | hsa00052 | Galactose metabolism | 1.194E-02 | 5.805E-02 | AKR1B10 |
| 09100 Metabolism | 09101 Carbohydrate metabolism | hsa00051 | Fructose and mannose metabolism | 1.273E-02 | 5.805E-02 | AKR1B10 |
| 09150 Organismal Systems | 09154 Digestive system | hsa04975 | Fat digestion and absorption | 1.629E-02 | 5.805E-02 | PLA2G1B |
| 09120 Genetic Information Processing | 09124 Replication and repair | hsa03410 | Base excision repair | 1.313E-02 | 5.805E-02 | POLB |
| 09100 Metabolism | 09103 Lipid metabolism | hsa00561 | Glycerolipid metabolism | 2.336E-02 | 6.332E-02 | AKR1B10 |
| 09100 Metabolism | 09103 Lipid metabolism | hsa00590 | Arachidonic acid metabolism | 2.454E-02 | 6.332E-02 | PLA2G1B |
| 09100 Metabolism | 09103 Lipid metabolism | hsa00565 | Ether lipid metabolism | 1.786E-02 | 5.805E-02 | PLA2G1B |
Human Diseases Associated with the Target Proteins of the Plant
Detailed Information of Human Diseases
| ICD10 Disease Category | Disease Name | ICD10 Code | Associated Disease Targets (Association Ref: TTD database) |
|---|---|---|---|
| C00-D49: Neoplasms | Cancer | C00-C96 | CDC25B; |


