Plant Name: Arisaema curvatum
Taxonomic Information:
Plant ID: NPO26204
Plant Latin Name: Arisaema curvatum
Taxonomy Genus: Arisaema
Taxonomy Family: Araceae
Plant External Links:
NCBI TaxonomyDB:
n.a.
Plant-of-the-World-Online:
n.a.
Collective Molecular Activities of the Plant
List of Human Proteins Collectively Targeted by the Plant
Click Gene ID to Access Activity Profile of Individual Target
List of Human Cytochrome P450 Enzymes Affected by the Plant
Click Gene ID to Access Activity Profile of Individual Enzyme
| Cytochrome P450 Enzymes: |
|---|
Detailed Information of Target Proteins
| Protein Class | Gene ID | Protein Name | Uniprot ID | Target ChEMBL ID |
|---|---|---|---|---|
| Hydrolase | BCHE | Butyrylcholinesterase | P06276 | CHEMBL1914 |
| Hydrolase | ACHE | Acetylcholinesterase | P22303 | CHEMBL220 |
| Plant homeodomain | KMT2A | Histone-lysine N-methyltransferase MLL | Q03164 | CHEMBL1293299 |
| Unclassified | MITF | Microphthalmia-associated transcription factor | O75030 | CHEMBL1741165 |
| Unclassified | PAX8 | Paired box protein Pax-8 | Q06710 | CHEMBL2362980 |
Enrichment of Gene Ontology of Target Proteins (Activity≤10μM)
Detailed Information of Enriched Gene Ontolgy
| GO Type | GO Category | Enriched GO Terms | p-Value | Adjusted p-Value | Enriched Genes |
|---|---|---|---|---|---|
| MF | GO:0003824; catalytic activity | GO:0003990; acetylcholinesterase activity | 5.361E-08 | 1.167E-03 | ACHE, BCHE |
| BP | GO:0032502; developmental process | GO:0048513; animal organ development | 6.816E-05 | 1.925E-01 | ACHE, KMT2A, MITF, PAX8 |
| MF | GO:0005488; binding | GO:0042165; neurotransmitter binding | 7.071E-05 | 1.925E-01 | ACHE, BCHE |
| MF | GO:0005488; binding | GO:0001540; amyloid-beta binding | 9.432E-05 | 2.054E-01 | ACHE, BCHE |
| BP | GO:0023052; signaling | GO:0050805; negative regulation of synaptic transmission | 1.074E-04 | 2.126E-01 | ACHE, BCHE |
| BP | GO:0008152; metabolic process | GO:0042133; neurotransmitter metabolic process | 2.034E-04 | 2.874E-01 | ACHE, BCHE |
| BP | GO:0009987; cellular process | GO:0038194; thyroid-stimulating hormone signaling pathway | 5.177E-04 | 2.874E-01 | PAX8 |
| MF | GO:0060089; molecular transducer activity | GO:0004996; thyroid-stimulating hormone receptor activity | 5.177E-04 | 2.874E-01 | PAX8 |
| MF | GO:0005488; binding | GO:1990837; sequence-specific double-stranded DNA binding | 5.412E-04 | 2.874E-01 | KMT2A, MITF, PAX8 |
| CC | GO:0044464; cell part | GO:0005641; nuclear envelope lumen | 2.069E-03 | 4.461E-01 | BCHE |
| CC | GO:0043226; organelle | GO:0005654; nucleoplasm | 2.977E-02 | 1.000E+00 | KMT2A, MITF, PAX8 |
Enriched KEGG Pathways of Target Proteins (Activity≤10μM)
Top 10 KEGG Pathways (ordered by adjusted p-value)
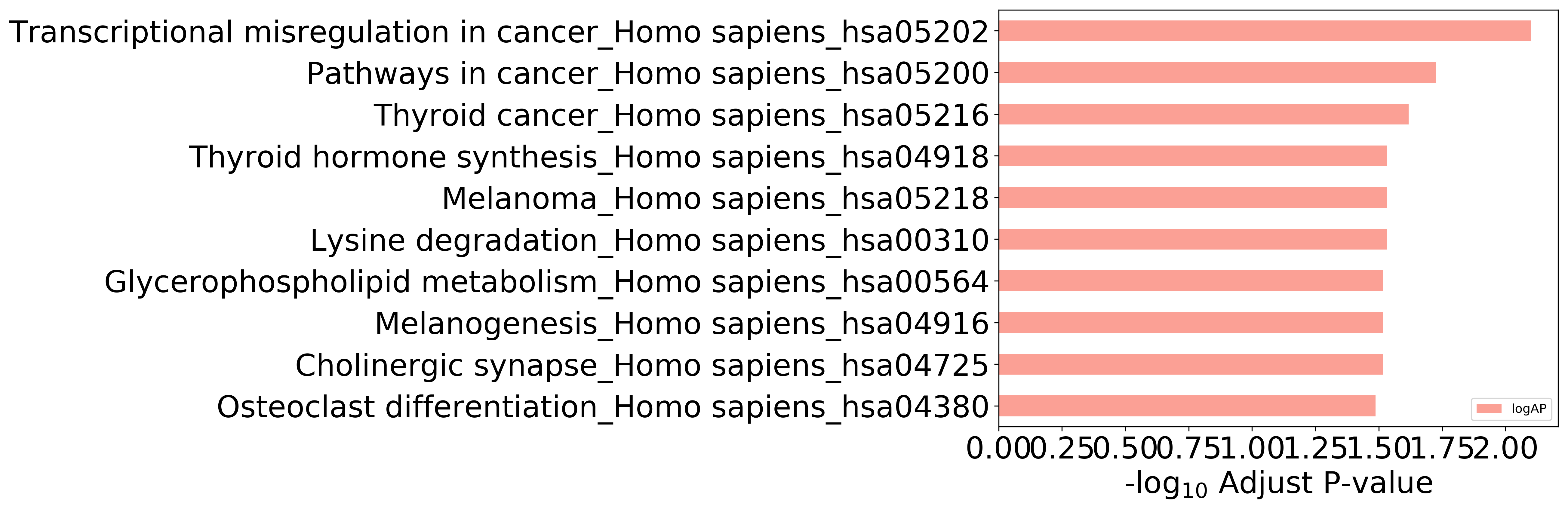
Detailed Information of Enriched KEGG Pathways
| Pathway Category Top Level | Pathway Category Second Level | Pathway ID | Pathway Name | p-Value | Adjusted p-Value | Enriched Genes |
|---|---|---|---|---|---|---|
| 09160 Human Diseases | 09161 Cancers | hsa05202 | Transcriptional misregulation in cancer | 7.913E-04 | 7.913E-03 | KMT2A, PAX8 |
| 09160 Human Diseases | 09161 Cancers | hsa05200 | Pathways in cancer | 3.778E-03 | 1.889E-02 | PAX8, MITF |
| 09160 Human Diseases | 09161 Cancers | hsa05216 | Thyroid cancer | 7.230E-03 | 2.410E-02 | PAX8 |
| 09100 Metabolism | 09105 Amino acid metabolism | hsa00310 | Lysine degradation | 1.293E-02 | 2.938E-02 | KMT2A |
| 09160 Human Diseases | 09161 Cancers | hsa05218 | Melanoma | 1.763E-02 | 2.938E-02 | MITF |
| 09150 Organismal Systems | 09152 Endocrine system | hsa04918 | Thyroid hormone synthesis | 1.763E-02 | 2.938E-02 | PAX8 |
| 09150 Organismal Systems | 09156 Nervous system | hsa04725 | Cholinergic synapse | 2.745E-02 | 3.050E-02 | ACHE |
| 09150 Organismal Systems | 09152 Endocrine system | hsa04916 | Melanogenesis | 2.475E-02 | 3.050E-02 | MITF |
| 09100 Metabolism | 09103 Lipid metabolism | hsa00564 | Glycerophospholipid metabolism | 2.353E-02 | 3.050E-02 | ACHE |
| 09150 Organismal Systems | 09158 Development | hsa04380 | Osteoclast differentiation | 3.257E-02 | 3.257E-02 | MITF |
Human Diseases Associated with the Target Proteins of the Plant
Detailed Information of Human Diseases
| ICD10 Disease Category | Disease Name | ICD10 Code | Associated Disease Targets (Association Ref: TTD database) |
|---|


