Plant Name: Anagallis arvensis
Taxonomic Information:
Plant ID: NPO18677
Plant Latin Name: Anagallis arvensis
Taxonomy Genus: Anagallis
Taxonomy Family: Primulaceae
Plant External Links:
NCBI TaxonomyDB:
4337
Plant-of-the-World-Online:
n.a.
Used in Medicines:
Country/Region:
Italy; China; India; SpainTraditional Medicine System:
Unani; TCM; HomeopathyMedicinal Functions:
Antidepressant; Antipruritic; Antitussive; Antiviral; Cholagogue; Diaphoretic; Diuretic; Expectorant; Homeopathy; Nervine; Purgative; Stimulant; Vulnerary
Geographical Distribution:
India; Italy; China; Spain
Example Image
Collective Molecular Activities of the Plant
List of Human Proteins Collectively Targeted by the Plant
Click Gene ID to Access Activity Profile of Individual Target
| PDE4D; | |
| CA1; CA7; CA12; CA9; CA13; | |
| RAB9A; NPC1; |
List of Human Cytochrome P450 Enzymes Affected by the Plant
Click Gene ID to Access Activity Profile of Individual Enzyme
| Cytochrome P450 Enzymes: |
|---|
Detailed Information of Target Proteins
| Protein Class | Gene ID | Protein Name | Uniprot ID | Target ChEMBL ID |
|---|---|---|---|---|
| Lyase | CA1 | Carbonic anhydrase I | P00915 | CHEMBL261 |
| Lyase | CA7 | Carbonic anhydrase VII | P43166 | CHEMBL2326 |
| Lyase | CA12 | Carbonic anhydrase XII | O43570 | CHEMBL3242 |
| Lyase | CA9 | Carbonic anhydrase IX | Q16790 | CHEMBL3594 |
| Lyase | CA13 | Carbonic anhydrase XIII | Q8N1Q1 | CHEMBL3912 |
| Phosphodiesterase 4 | PDE4D | Phosphodiesterase 4D | Q08499 | CHEMBL288 |
| Unclassified | RAB9A | Ras-related protein Rab-9A | P51151 | CHEMBL1293294 |
| Unclassified | NPC1 | Niemann-Pick C1 protein | O15118 | CHEMBL1293277 |
Enrichment of Gene Ontology of Target Proteins (Activity≤10μM)
Detailed Information of Enriched Gene Ontolgy
| GO Type | GO Category | Enriched GO Terms | p-Value | Adjusted p-Value | Enriched Genes |
|---|---|---|---|---|---|
| MF | GO:0003824; catalytic activity | GO:0004089; carbonate dehydratase activity | 7.499E-15 | 1.633E-10 | CA1, CA12, CA13, CA7, CA9 |
| BP | GO:0008152; metabolic process | GO:0006730; one-carbon metabolic process | 9.388E-13 | 1.022E-08 | CA1, CA12, CA13, CA7, CA9 |
| BP | GO:0051179; localization | GO:0015701; bicarbonate transport | 2.702E-12 | 1.961E-08 | CA1, CA12, CA13, CA7, CA9 |
| MF | GO:0005488; binding | GO:0008270; zinc ion binding | 6.573E-06 | 1.301E-02 | CA1, CA12, CA13, CA7, CA9 |
| CC | GO:0044464; cell part | GO:0005764; lysosome | 5.306E-03 | 1.000E+00 | NPC1, RAB9A |
| CC | GO:0043226; organelle | GO:0042470; melanosome | 4.108E-02 | 1.000E+00 | RAB9A |
| MF | GO:0005488; binding | GO:0005496; steroid binding | 3.507E-02 | 1.000E+00 | NPC1 |
| MF | GO:0003824; catalytic activity | GO:0008081; phosphoric diester hydrolase activity | 3.708E-02 | 1.000E+00 | PDE4D |
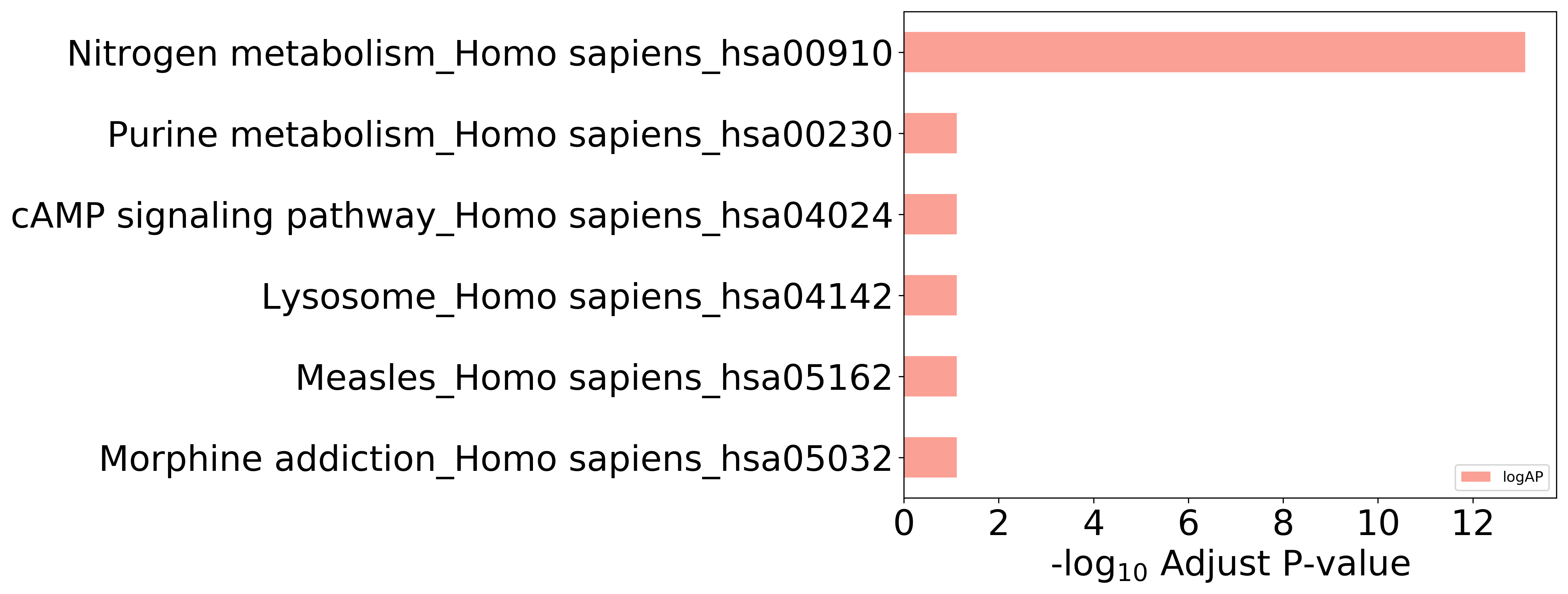
Enriched KEGG Pathways of Target Proteins (Activity≤10μM)
Top 10 KEGG Pathways (ordered by adjusted p-value)
Detailed Information of Enriched KEGG Pathways
| Pathway Category Top Level | Pathway Category Second Level | Pathway ID | Pathway Name | p-Value | Adjusted p-Value | Enriched Genes |
|---|---|---|---|---|---|---|
| 09100 Metabolism | 09102 Energy metabolism | hsa00910 | Nitrogen metabolism | 1.298E-14 | 7.789E-14 | CA12, CA1, CA7, CA9, CA13 |
| 09160 Human Diseases | 09164 Substance dependence | hsa05032 | Morphine addiction | 3.583E-02 | 7.690E-02 | PDE4D |
| 09160 Human Diseases | 09167 Infectious diseases | hsa05162 | Measles | 5.313E-02 | 7.690E-02 | RAB9A |
| 09140 Cellular Processes | 09141 Transport and catabolism | hsa04142 | Lysosome | 4.816E-02 | 7.690E-02 | NPC1 |
| 09130 Environmental Information Processing | 09132 Signal transduction | hsa04024 | cAMP signaling pathway | 7.690E-02 | 7.690E-02 | PDE4D |
| 09100 Metabolism | 09104 Nucleotide metabolism | hsa00230 | Purine metabolism | 6.828E-02 | 7.690E-02 | PDE4D |
Human Diseases Associated with the Target Proteins of the Plant
Detailed Information of Human Diseases
| ICD10 Disease Category | Disease Name | ICD10 Code | Associated Disease Targets (Association Ref: TTD database) |
|---|---|---|---|
| C00-D49: Neoplasms | Lymphoma | C81-C86 | CA9; |
| C00-D49: Neoplasms | Cancer | C00-C96 | CA9; |
| C00-D49: Neoplasms | Clear cell renal cell carcinoma | C64 | CA9; |
| C00-D49: Neoplasms | Solid tumours | C00-D48 | CA9; |
| C00-D49: Neoplasms | Breast cancer | C50 | CA9; |
| C00-D49: Neoplasms | Renal cancer | C64 | CA9; |


