Plant Name: Cajanus cajan
Taxonomic Information:
Plant ID: NPO18252
Plant Latin Name: Cajanus cajan
Taxonomy Genus: Cajanus
Taxonomy Family: Fabaceae
Plant External Links:
NCBI TaxonomyDB:
3821
Plant-of-the-World-Online:
1152177-2
Used in Medicines:
Country/Region:
Brazil; Madagascar; Angola; South Africa; Tanzania; Indonesia; India; Zimbabwe; Malaysia; Vietnam; Rwanda; Swaziland; Laos; Kenya; Nigeria; ThailandTraditional Medicine System:
Sowa-Rigpa; Indian Folk; Unani; Ayurveda; SiddhaGeographical Distribution:
Turkmenistan; Cambodia; Ethiopia; Swaziland; Bolivia; Cameroon; Burkina Faso; Ghana; Saudi Arabia; Cape Verde; Liberia; Paraguay; Jamaica; Tanzania; French Guiana; Yemen; Pakistan; India; Kenya; Tajikistan; Afghanistan; Bangladesh; Rwanda; Somalia; Peru; Laos; Cote d'Ivoire; Benin; Cuba; Togo; China; Dominican Republic; Indonesia; United States; Vietnam; Mali; Russia; Mauritius; Angola; Chad; South Africa; Malaysia; Senegal; Mozambique; Uganda; Niger; Brazil; Guinea; Bahamas; Nigeria; Ecuador; Australia; Thailand; Haiti; Sierra Leone; Gambia; Philippines; Namibia; Guinea-Bissau; Seychelles; Uzbekistan; Djibouti; Colombia; Burundi; Taiwan; Madagascar; Sudan; Nepal; Suriname; Venezuela; Zambia; Papua New Guinea; Malawi; Zimbabwe; Eritrea; New Caledonia; Trinidad and Tobago; Guyana; Honduras; Myanmar; Mexico; Egypt; Congo; Sri Lanka; Gabon; Comoros
Example Image

Collective Molecular Activities of the Plant
List of Human Proteins Collectively Targeted by the Plant
Click Gene ID to Access Activity Profile of Individual Target
| NR1H4; | |
| ELANE; | |
| MAPT; |
List of Human Cytochrome P450 Enzymes Affected by the Plant
Click Gene ID to Access Activity Profile of Individual Enzyme
| Cytochrome P450 Enzymes: |
|---|
Detailed Information of Target Proteins
| Protein Class | Gene ID | Protein Name | Uniprot ID | Target ChEMBL ID |
|---|---|---|---|---|
| Nuclear hormone receptor subfamily 1 group H | NR1H4 | Bile acid receptor FXR | Q96RI1 | CHEMBL2047 |
| Serine protease | ELANE | Leukocyte elastase | P08246 | CHEMBL248 |
| Unclassified | MAPT | Microtubule-associated protein tau | P10636 | CHEMBL1293224 |
Enrichment of Gene Ontology of Target Proteins (Activity≤10μM)
Detailed Information of Enriched Gene Ontolgy
| GO Type | GO Category | Enriched GO Terms | p-Value | Adjusted p-Value | Enriched Genes |
|---|---|---|---|---|---|
| BP | GO:0032501; multicellular organismal process | GO:0032682; negative regulation of chemokine production | 2.459E-06 | 5.355E-02 | ELANE, NR1H4 |
| BP | GO:0050896; response to stimulus | GO:0050728; negative regulation of inflammatory response | 1.050E-04 | 1.353E-01 | ELANE, NR1H4 |
| MF | GO:0140110; transcription regulator activity | GO:0001076; transcription factor activity, RNA polymerase II transcription factor binding | 1.909E-04 | 1.485E-01 | ELANE, NR1H4 |
| MF | GO:0060089; molecular transducer activity | GO:0038181; bile acid receptor activity | 4.659E-04 | 2.359E-01 | NR1H4 |
| MF | GO:0060089; molecular transducer activity | GO:0004887; thyroid hormone receptor activity | 1.087E-03 | 3.242E-01 | NR1H4 |
| MF | GO:0005488; binding | GO:0034452; dynactin binding | 1.708E-03 | 3.610E-01 | MAPT |
| MF | GO:0005488; binding | GO:0046965; retinoid X receptor binding | 2.328E-03 | 4.121E-01 | NR1H4 |
| CC | GO:0044464; cell part | GO:0030673; axolemma | 2.483E-03 | 4.191E-01 | MAPT |
Enriched KEGG Pathways of Target Proteins (Activity≤10μM)
Top 10 KEGG Pathways (ordered by adjusted p-value)
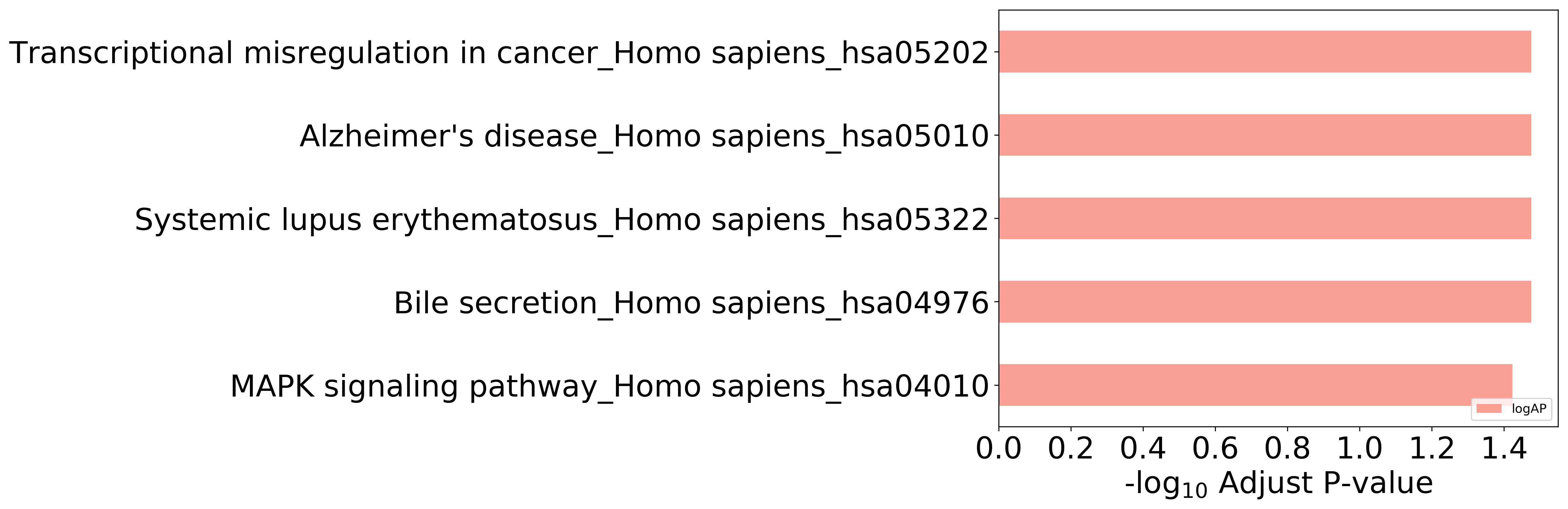
Detailed Information of Enriched KEGG Pathways
| Pathway Category Top Level | Pathway Category Second Level | Pathway ID | Pathway Name | p-Value | Adjusted p-Value | Enriched Genes |
|---|---|---|---|---|---|---|
| 09150 Organismal Systems | 09154 Digestive system | hsa04976 | Bile secretion | 1.061E-02 | 3.345E-02 | NR1H4 |
| 09160 Human Diseases | 09162 Immune diseases | hsa05322 | Systemic lupus erythematosus | 2.011E-02 | 3.345E-02 | ELANE |
| 09160 Human Diseases | 09163 Neurodegenerative diseases | hsa05010 | Alzheimer's disease | 2.499E-02 | 3.345E-02 | MAPT |
| 09130 Environmental Information Processing | 09132 Signal transduction | hsa04010 | MAPK signaling pathway | 3.777E-02 | 3.777E-02 | MAPT |
| 09160 Human Diseases | 09161 Cancers | hsa05202 | Transcriptional misregulation in cancer | 2.676E-02 | 3.345E-02 | ELANE |
Human Diseases Associated with the Target Proteins of the Plant
Detailed Information of Human Diseases
| ICD10 Disease Category | Disease Name | ICD10 Code | Associated Disease Targets (Association Ref: TTD database) |
|---|


