Plant Name: Euphorbia royleana
Taxonomic Information:
Plant ID: NPO12595
Plant Latin Name: Euphorbia royleana
Taxonomy Genus: Euphorbia
Taxonomy Family: Euphorbiaceae
Plant External Links:
NCBI TaxonomyDB:
1130020
Plant-of-the-World-Online:
n.a.
Used in Medicines:
Country/Region:
China; IndiaTraditional Medicine System:
TCM; Unani; AyurvedaGeographical Distribution:
India; China
Collective Molecular Activities of the Plant
List of Human Proteins Collectively Targeted by the Plant
Click Gene ID to Access Activity Profile of Individual Target
| CA7; CA9; CA12; | |
| PPARA; PPARD; | |
| PTGS1; | |
| GFER; ALDH1A1; HSD17B10; IDO1; | |
| CASP7; CASP1; | |
| TLR2; | |
| ABCB1; | |
| FABP3; LMNA; FABP2; FABP4; FABP5; |
List of Human Cytochrome P450 Enzymes Affected by the Plant
Click Gene ID to Access Activity Profile of Individual Enzyme
| Cytochrome P450 Enzymes: | CYP1A2; |
|---|
Detailed Information of Target Proteins
| Protein Class | Gene ID | Protein Name | Uniprot ID | Target ChEMBL ID |
|---|---|---|---|---|
| ATP-binding cassette | ABCB1 | P-glycoprotein 1 | P08183 | CHEMBL4302 |
| Cysteine protease | CASP7 | Caspase-7 | P55210 | CHEMBL3468 |
| Cysteine protease | CASP1 | Caspase-1 | P29466 | CHEMBL4801 |
| Cytochrome P450 family 1 | CYP1A2 | Cytochrome P450 1A2 | P05177 | CHEMBL3356 |
| Enzyme_unclassified | GFER | FAD-linked sulfhydryl oxidase ALR | P55789 | CHEMBL1741189 |
| Enzyme_unclassified | ALDH1A1 | Aldehyde dehydrogenase 1A1 | P00352 | CHEMBL3577 |
| Enzyme_unclassified | HSD17B10 | Endoplasmic reticulum-associated amyloid beta-peptide-binding protein | Q99714 | CHEMBL4159 |
| Enzyme_unclassified | IDO1 | Indoleamine 2,3-dioxygenase | P14902 | CHEMBL4685 |
| Lyase | CA7 | Carbonic anhydrase VII | P43166 | CHEMBL2326 |
| Lyase | CA9 | Carbonic anhydrase IX | Q16790 | CHEMBL3594 |
| Lyase | CA12 | Carbonic anhydrase XII | O43570 | CHEMBL3242 |
| Nuclear hormone receptor subfamily 1 group C | PPARA | Peroxisome proliferator-activated receptor alpha | Q07869 | CHEMBL239 |
| Nuclear hormone receptor subfamily 1 group C | PPARD | Peroxisome proliferator-activated receptor delta | Q03181 | CHEMBL3979 |
| Oxidoreductase | PTGS1 | Cyclooxygenase-1 | P23219 | CHEMBL221 |
| Toll-like and Il-1 receptors | TLR2 | Toll-like receptor 2 | O60603 | CHEMBL4163 |
| Unclassified | FABP3 | Fatty acid binding protein muscle | P05413 | CHEMBL3344 |
| Unclassified | LMNA | Prelamin-A/C | P02545 | CHEMBL1293235 |
| Unclassified | FABP2 | Fatty acid binding protein intestinal | P12104 | CHEMBL4879 |
| Unclassified | FABP4 | Fatty acid binding protein adipocyte | P15090 | CHEMBL2083 |
| Unclassified | FABP5 | Fatty acid binding protein epidermal | Q01469 | CHEMBL3674 |
Enrichment of Gene Ontology of Target Proteins (Activity≤10μM)
Detailed Information of Enriched Gene Ontolgy
| GO Type | GO Category | Enriched GO Terms | p-Value | Adjusted p-Value | Enriched Genes |
|---|---|---|---|---|---|
| BP | GO:0008152; metabolic process | GO:0019433; triglyceride catabolic process | 1.042E-08 | 7.566E-05 | FABP2, FABP3, FABP4, FABP5 |
| MF | GO:0003824; catalytic activity | GO:0004089; carbonate dehydratase activity | 4.285E-07 | 7.775E-04 | CA12, CA7, CA9 |
| BP | GO:0032501; multicellular organismal process | GO:0001816; cytokine production | 1.457E-06 | 1.586E-03 | CASP1, FABP4, IDO1, TLR2 |
| BP | GO:0050896; response to stimulus | GO:0050727; regulation of inflammatory response | 1.930E-06 | 1.827E-03 | CASP1, FABP4, IDO1, PPARA, PPARD, TLR2 |
| BP | GO:0050896; response to stimulus | GO:0033993; response to lipid | 5.014E-06 | 4.043E-03 | CA9, CASP1, CYP1A2, FABP3, IDO1, PPARD, TLR2 |
| BP | GO:0050896; response to stimulus | GO:0001666; response to hypoxia | 5.483E-06 | 4.264E-03 | CASP1, LMNA, PPARA, PPARD, TLR2 |
| BP | GO:0008152; metabolic process | GO:0006730; one-carbon metabolic process | 6.631E-06 | 4.658E-03 | CA12, CA7, CA9 |
| BP | GO:0008152; metabolic process | GO:0006631; fatty acid metabolic process | 1.049E-05 | 6.343E-03 | CYP1A2, FABP3, PPARA, PPARD, PTGS1 |
| BP | GO:0051704; multi-organism process | GO:0002237; response to molecule of bacterial origin | 1.049E-05 | 6.343E-03 | CASP1, CYP1A2, IDO1, PPARD, TLR2 |
| BP | GO:0051179; localization | GO:0015701; bicarbonate transport | 1.224E-05 | 7.201E-03 | CA12, CA7, CA9 |
| MF | GO:0005488; binding | GO:0036041; long-chain fatty acid binding | 5.571E-05 | 2.696E-02 | FABP3, PPARD |
| MF | GO:0003824; catalytic activity | GO:0016491; oxidoreductase activity | 7.022E-05 | 3.186E-02 | ALDH1A1, CYP1A2, GFER, HSD17B10, IDO1, PTGS1 |
| MF | GO:0003824; catalytic activity | GO:0097200; cysteine-type endopeptidase activity involved in execution phase of apoptosis | 7.890E-05 | 3.506E-02 | CASP1, CASP7 |
| MF | GO:0005488; binding | GO:0048037; cofactor binding | 1.088E-04 | 4.387E-02 | ALDH1A1, CYP1A2, GFER, IDO1, PTGS1 |
Enriched KEGG Pathways of Target Proteins (Activity≤10μM)
Top 10 KEGG Pathways (ordered by adjusted p-value)
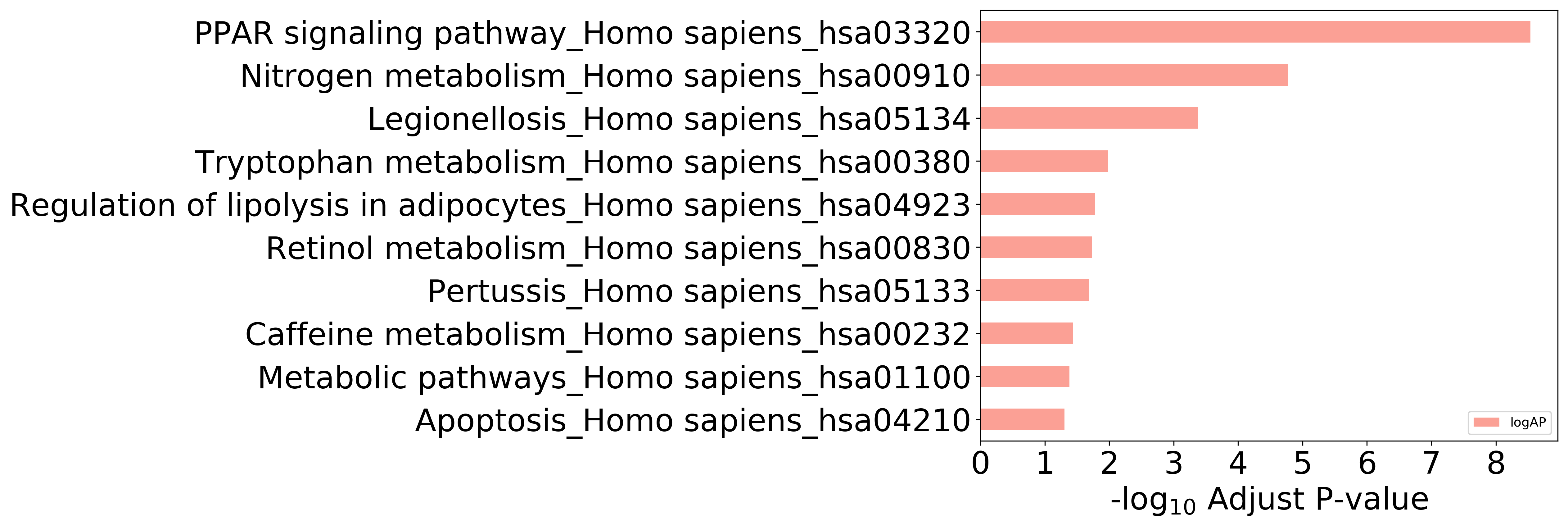
Detailed Information of Enriched KEGG Pathways
| Pathway Category Top Level | Pathway Category Second Level | Pathway ID | Pathway Name | p-Value | Adjusted p-Value | Enriched Genes |
|---|---|---|---|---|---|---|
| 09150 Organismal Systems | 09152 Endocrine system | hsa03320 | PPAR signaling pathway | 5.037E-11 | 2.921E-09 | FABP2, FABP3, FABP4, FABP5, PPARA, PPARD |
| 09100 Metabolism | 09102 Energy metabolism | hsa00910 | Nitrogen metabolism | 5.763E-07 | 1.671E-05 | CA12, CA7, CA9 |
| 09160 Human Diseases | 09167 Infectious diseases | hsa05134 | Legionellosis | 2.170E-05 | 4.196E-04 | CASP7, CASP1, TLR2 |
| 09100 Metabolism | 09105 Amino acid metabolism | hsa00380 | Tryptophan metabolism | 7.243E-04 | 1.050E-02 | CYP1A2, IDO1 |
| 09150 Organismal Systems | 09152 Endocrine system | hsa04923 | Regulation of lipolysis in adipocytes | 1.416E-03 | 1.643E-02 | FABP4, PTGS1 |
| 09100 Metabolism | 09108 Metabolism of cofactors and vitamins | hsa00830 | Retinol metabolism | 1.903E-03 | 1.839E-02 | CYP1A2, ALDH1A1 |
| 09160 Human Diseases | 09167 Infectious diseases | hsa05133 | Pertussis | 2.524E-03 | 2.091E-02 | CASP7, CASP1 |
| Unclassified | Unclassified | hsa01100 | Metabolic pathways | 6.417E-03 | 4.135E-02 | CYP1A2, ALDH1A1, HSD17B10, IDO1, PTGS1 |
| 09140 Cellular Processes | 09143 Cell growth and death | hsa04210 | Apoptosis | 8.511E-03 | 4.936E-02 | CASP7, LMNA |
| 09100 Metabolism | 09110 Biosynthesis of other secondary metabolites | hsa00232 | Caffeine metabolism | 4.991E-03 | 3.618E-02 | CYP1A2 |
Human Diseases Associated with the Target Proteins of the Plant
Detailed Information of Human Diseases
| ICD10 Disease Category | Disease Name | ICD10 Code | Associated Disease Targets (Association Ref: TTD database) |
|---|---|---|---|
| C00-D49: Neoplasms | Clear cell renal cell carcinoma | C64 | CA9; |
| C00-D49: Neoplasms | Cancer | C00-C96 | CA9; |
| C00-D49: Neoplasms | Lymphoma | C81-C86 | CA9; |
| C00-D49: Neoplasms | Renal cancer | C64 | CA9; |
| C00-D49: Neoplasms | Solid tumours | C00-D48 | CA9; |
| C00-D49: Neoplasms | Breast cancer | C50 | CA9; |


