Plant Name: Stachyurus himalaicus
Collective Molecular Activities at 10μM Activity Cutoff
List of Human Proteins Collectively Targeted by the Plant
Click Gene ID to Access Activity Profile of Individual Target
| ADORA3; ADORA2A; ADORA1; | |
| TSHR; | |
| RECQL; TDP1; ADK; PKM; GLO1; ALOX12; HSD17B1; AKR1B1; HSD17B10; NOX4; ALOX15; USP2; TNKS2; TNKS; PARP1; CD38; POLB; | |
| ACHE; | |
| CFTR; | |
| GSK3B; SYK; FLT3; CDK6; CSNK2A1; | |
| CA2; CA12; CA7; CA4; | |
| AR; | |
| ESR2; ESR1; | |
| NR1H4; | |
| ALOX5; MAOA; PTGS2; XDH; | |
| KDM4E; | |
| MMP9; MMP12; MMP2; | |
| STAT6; TP53; HIF1A; | |
| ABCG2; ABCC1; | |
| SLC28A2; SLC28A3; SLC28A1; SLCO1B3; SLCO1B1; | |
| LMNA; NPC1; THPO; MAPT; APP; TTR; | |
| SMN1;SMN2; |
List of Human Cytochrome P450 Enzymes Affected by the Plant
Click Gene ID to Access Activity Profile of Individual Enzyme
Detailed Information of Human Protein Targets
| Protein Class | Gene ID | Protein Name | Uniprot ID | Target ChEMBL ID |
|---|---|---|---|---|
| ATP-binding cassette | ABCG2 | ATP-binding cassette sub-family G member 2 | Q9UNQ0 | CHEMBL5393 |
| ATP-binding cassette | ABCC1 | Multidrug resistance-associated protein 1 | P33527 | CHEMBL3004 |
| Chloride channel | CFTR | Cystic fibrosis transmembrane conductance regulator | P13569 | CHEMBL4051 |
| Cytochrome P450 family 1 | CYP1A2 | Cytochrome P450 1A2 | P05177 | CHEMBL3356 |
| Cytochrome P450 family 1 | CYP1B1 | Cytochrome P450 1B1 | Q16678 | CHEMBL4878 |
| Cytochrome P450 family 1 | CYP1A1 | Cytochrome P450 1A1 | P04798 | CHEMBL2231 |
| Cytochrome P450 family 19 | CYP19A1 | Cytochrome P450 19A1 | P11511 | CHEMBL1978 |
| Cytochrome P450 family 3 | CYP3A4 | Cytochrome P450 3A4 | P08684 | CHEMBL340 |
| Enzyme_unclassified | ALOX15 | Arachidonate 15-lipoxygenase | P16050 | CHEMBL2903 |
| Enzyme_unclassified | USP2 | Ubiquitin carboxyl-terminal hydrolase 2 | O75604 | CHEMBL1293227 |
Showing 1 to 10 of 66 entries
Enrichment of Gene Ontology of Target Proteins (Activity≤10μM)
Detailed Information of Enriched Gene Ontolgy
| GO Type | GO Category | Enriched GO Terms | p-Value | Adjusted p-Value | Enriched Genes |
|---|---|---|---|---|---|
| BP | Unclassified; | GO:0032844; regulation of homeostatic process | 6.253E-11 | 8.510E-08 | ADORA1, ADORA2A, CA2, CA7, CD38, CDK6, CFTR, HIF1A, NPC1, NR1H4, PARP1, PTGS2, SYK, TNKS, TNKS2 |
| BP | Unclassified; | GO:0032846; positive regulation of homeostatic process | 3.188E-10 | 3.155E-07 | ADORA2A, CA2, CA7, CFTR, HIF1A, NPC1, NR1H4, PTGS2, SYK, TNKS, TNKS2 |
| BP | GO:0008152; metabolic process | GO:0006691; leukotriene metabolic process | 6.834E-08 | 1.958E-05 | ABCC1, ALOX12, ALOX15, ALOX5, SYK |
| BP | GO:0008152; metabolic process | GO:0019372; lipoxygenase pathway | 2.332E-07 | 5.288E-05 | ALOX12, ALOX15, ALOX5, PTGS2 |
| BP | GO:0051179; localization | GO:0015701; bicarbonate transport | 4.219E-07 | 8.506E-05 | CA12, CA2, CA4, CA7, CFTR |
| BP | GO:0008152; metabolic process | GO:0016579; protein deubiquitination | 5.737E-07 | 1.120E-04 | AR, CFTR, ESR1, HIF1A, POLB, TNKS, TNKS2, TP53, USP2 |
| BP | GO:0032501; multicellular organismal process | GO:0048143; astrocyte activation | 3.301E-06 | 5.171E-04 | ADORA2A, APP, MAPT |
| BP | GO:0009987; cellular process | GO:0001973; adenosine receptor signaling pathway | 3.301E-06 | 5.171E-04 | ADORA1, ADORA2A, ADORA3 |
| BP | GO:0051179; localization | GO:2001225; regulation of chloride transport | 4.704E-06 | 6.829E-04 | CA2, CA7, CFTR |
| BP | GO:0008152; metabolic process | GO:0097267; omega-hydroxylase P450 pathway | 4.704E-06 | 6.829E-04 | CYP1A1, CYP1A2, CYP1B1 |
Showing 1 to 10 of 87 entries
Enriched KEGG Pathways of Target Proteins (Activity≤10μM)
Top 10, ordered by adjusted p-value
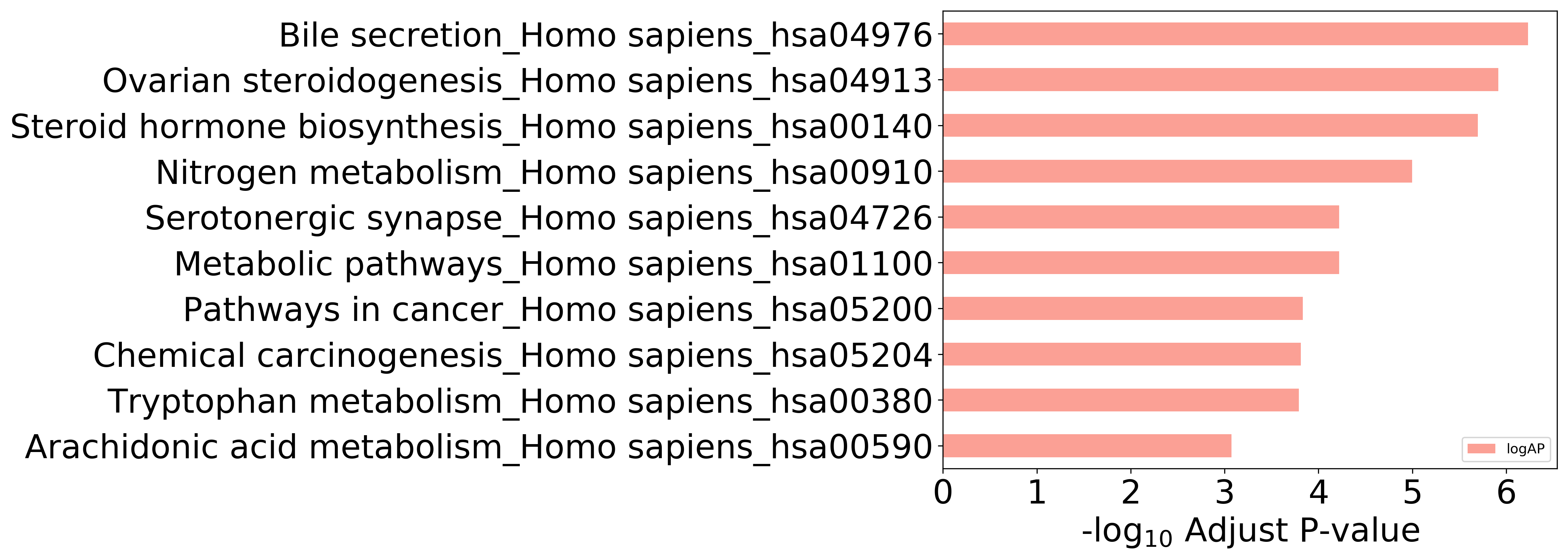
Detailed Information of Enriched KEGG Pathways
| Pathway ID | Pathway Name | p-Value | Adjusted p-Value | Enriched Genes |
|---|---|---|---|---|
| hsa00140 | Steroid hormone biosynthesis_Homo sapiens_hsa00140 | 3.969E-08 | 2.011E-06 | HSD17B1; CYP1A2; CYP1A1; CYP1B1; CYP3A4; CYP19A1 |
| hsa00232 | Caffeine metabolism_Homo sapiens_hsa00232 | 1.098E-04 | 1.284E-03 | CYP1A2; XDH |
| hsa00380 | Tryptophan metabolism_Homo sapiens_hsa00380 | 9.599E-06 | 1.621E-04 | MAOA; CYP1A2; CYP1A1; CYP1B1 |
| hsa00590 | Arachidonic acid metabolism_Homo sapiens_hsa00590 | 5.544E-05 | 8.426E-04 | ALOX5; ALOX15; ALOX12; PTGS2 |
| hsa00591 | Linoleic acid metabolism_Homo sapiens_hsa00591 | 1.234E-04 | 1.339E-03 | CYP1A2; ALOX15; CYP3A4 |
| hsa00620 | Pyruvate metabolism_Homo sapiens_hsa00620 | 7.944E-03 | 3.096E-02 | PKM; GLO1 |
| hsa00830 | Retinol metabolism_Homo sapiens_hsa00830 | 1.353E-03 | 7.617E-03 | CYP1A2; CYP1A1; CYP3A4 |
| hsa00910 | Nitrogen metabolism_Homo sapiens_hsa00910 | 2.649E-07 | 1.007E-05 | CA12; CA2; CA4; CA7 |
| hsa00980 | Metabolism of xenobiotics by cytochrome P450_Homo sapiens_hsa00980 | 1.052E-04 | 1.284E-03 | CYP1A2; CYP1A1; CYP1B1; CYP3A4 |
| hsa00982 | Drug metabolism - cytochrome P450_Homo sapiens_hsa00982 | 1.608E-03 | 8.426E-03 | MAOA; CYP1A2; CYP3A4 |
Showing 1 to 10 of 47 entries
Human Diseases Associated with the Target Proteins of the Plant (Activity≤10μM)
Detailed Information of Human Diseases
| ICD10 Disease Category | Disease Name | ICD10 Code | Linked Targets |
|---|---|---|---|
| A00-B99: Certain infectious and parasitic diseases | Lymphatic filariasis | B74 | ALOX5 |
| A00-B99: Certain infectious and parasitic diseases | Diarrhea | A09, K59.1 | CFTR |
| A00-B99: Certain infectious and parasitic diseases | Pediculus humanus capitis | B85.0 | ACHE |
| A00-B99: Certain infectious and parasitic diseases | HIV-associated diarrhoea | A09 | CFTR |
| A00-B99: Certain infectious and parasitic diseases | Human immunodeficiency virus infection | B20-B26 | FLT3 |
| A00-B99: Certain infectious and parasitic diseases | Helminth infection | A00-B99 | ACHE |
| A00-B99: Certain infectious and parasitic diseases | Trematode infection | B66.9 | ESR1 |
| A00-B99: Certain infectious and parasitic diseases | Sepsis | A40, A41 | ADORA1 |
| A00-B99: Certain infectious and parasitic diseases | Malaria | B54 | ADORA3 |
| A00-B99: Certain infectious and parasitic diseases | Fungal infections | B35-B49 | ALOX5 |
Showing 1 to 10 of 222 entries