Plant Name: Tipuana tipu
Collective Molecular Activities at 10μM Activity Cutoff
List of Human Proteins Collectively Targeted by the Plant
Click Gene ID to Access Activity Profile of Individual Target
| TSHR; NPSR1; CXCR1; NMUR2; | |
| ADRA2C; ADRA2A; DRD4; | |
| RECQL; TDP1; PLA2G1B; ALPI; CTDSP1; PIK3R1; GLO1; MPO; HSD17B1; AKR1B1; HSD17B2; HSD17B10; ALOX15; ALOX12; NOX4; PYGL; POLB; | |
| ACHE; GAA; | |
| PKN1; AKT1; MET; CAMK2B; CSNK2A1; DAPK1; NEK2; FLT3; NEK6; SRC; IGF1R; PTK2; AURKB; GSK3B; CDK1; MAPK1; PIM1; NUAK1; ALK; AXL; KDR; | |
| CA14; CA3; CA7; CA5A; CA1; CA9; CA4; CA6; CA2; CA12; | |
| PPARA; PPARD; | |
| AR; | |
| ESRRA; | |
| KDM4E; | |
| MAOA; ALOX5; TYR; XDH; | |
| MMP13; MMP3; MMP2; MMP9; | |
| BACE1; | |
| AHR; STAT6; HIF1A; NFKB1; TP53; | |
| ABCC1; ABCG2; | |
| LMNA; MAPT; |
List of Human Cytochrome P450 Enzymes Affected by the Plant
Click Gene ID to Access Activity Profile of Individual Enzyme
Detailed Information of Human Protein Targets
| Protein Class | Gene ID | Protein Name | Uniprot ID | Target ChEMBL ID |
|---|---|---|---|---|
| Aspartic protease | BACE1 | Beta-secretase 1 | P56817 | CHEMBL4822 |
| ATP-binding cassette | ABCC1 | Multidrug resistance-associated protein 1 | P33527 | CHEMBL3004 |
| ATP-binding cassette | ABCG2 | ATP-binding cassette sub-family G member 2 | Q9UNQ0 | CHEMBL5393 |
| Cytochrome P450 family 1 | CYP1B1 | Cytochrome P450 1B1 | Q16678 | CHEMBL4878 |
| Cytochrome P450 family 1 | CYP1A1 | Cytochrome P450 1A1 | P04798 | CHEMBL2231 |
| Cytochrome P450 family 1 | CYP1A2 | Cytochrome P450 1A2 | P05177 | CHEMBL3356 |
| Cytochrome P450 family 19 | CYP19A1 | Cytochrome P450 19A1 | P11511 | CHEMBL1978 |
| Cytochrome P450 family 2 | CYP2C19 | Cytochrome P450 2C19 | P33261 | CHEMBL3622 |
| Cytochrome P450 family 2 | CYP2C9 | Cytochrome P450 2C9 | P11712 | CHEMBL3397 |
| Cytochrome P450 family 2 | CYP2D6 | Cytochrome P450 2D6 | P10635 | CHEMBL289 |
Showing 1 to 10 of 89 entries
Enrichment of Gene Ontology of Target Proteins (Activity≤10μM)
Detailed Information of Enriched Gene Ontolgy
| GO Type | GO Category | Enriched GO Terms | p-Value | Adjusted p-Value | Enriched Genes |
|---|---|---|---|---|---|
| BP | GO:0008152; metabolic process | GO:0097267; omega-hydroxylase P450 pathway | 1.668E-12 | 1.038E-09 | CYP1A1, CYP1A2, CYP1B1, CYP2C19, CYP2C8, CYP2C9 |
| BP | GO:0008152; metabolic process | GO:0016098; monoterpenoid metabolic process | 1.108E-11 | 5.026E-09 | CYP1A2, CYP2C19, CYP2C9, CYP2D6, CYP3A4 |
| BP | GO:0008152; metabolic process | GO:0019373; epoxygenase P450 pathway | 4.139E-10 | 1.146E-07 | CYP1A1, CYP1A2, CYP1B1, CYP2C19, CYP2C8, CYP2C9 |
| BP | GO:0050896; response to stimulus | GO:0042738; exogenous drug catabolic process | 1.431E-09 | 3.246E-07 | CYP1A2, CYP2C19, CYP2C8, CYP2C9, CYP3A4 |
| BP | GO:0008152; metabolic process | GO:0070989; oxidative demethylation | 1.431E-09 | 3.246E-07 | CYP1A2, CYP2C8, CYP2C9, CYP2D6, CYP3A4 |
| BP | GO:0008152; metabolic process | GO:0018105; peptidyl-serine phosphorylation | 6.893E-08 | 9.152E-06 | AKT1, AURKB, CAMK2B, CDK1, GSK3B, MAPK1, NEK6, PKN1, SRC |
| BP | GO:0008152; metabolic process | GO:0018107; peptidyl-threonine phosphorylation | 7.029E-08 | 9.232E-06 | AKT1, CAMK2B, CDK1, CSNK2A1, GSK3B, MAPK1, PKN1 |
| BP | GO:0008152; metabolic process | GO:0006691; leukotriene metabolic process | 2.865E-07 | 3.267E-05 | ABCC1, ALOX12, ALOX15, ALOX5, PLA2G1B |
| BP | GO:0009987; cellular process | GO:0030168; platelet activation | 7.517E-07 | 7.794E-05 | ADRA2A, ADRA2C, AKT1, AXL, MAPK1, PIK3R1, SRC |
| BP | GO:0008152; metabolic process | GO:0002933; lipid hydroxylation | 1.872E-06 | 1.773E-04 | CYP1A1, CYP2C8, CYP3A4 |
Showing 1 to 10 of 93 entries
Enriched KEGG Pathways of Target Proteins (Activity≤10μM)
Top 10, ordered by adjusted p-value
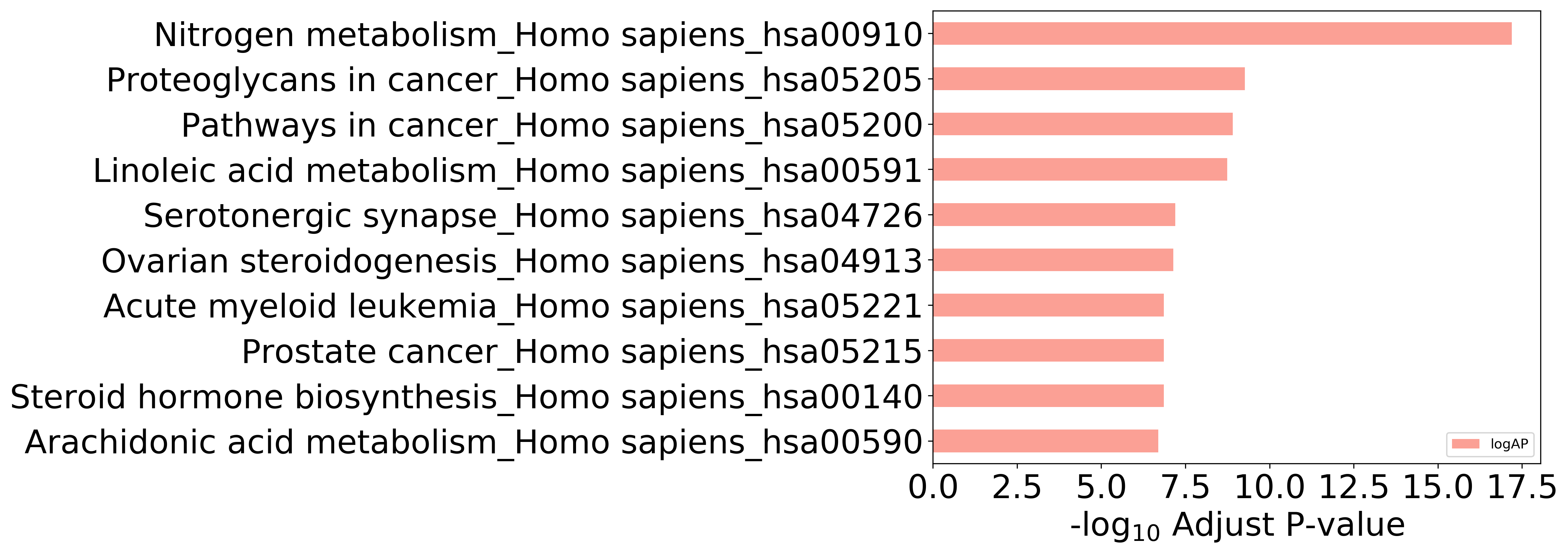
Detailed Information of Enriched KEGG Pathways
| Pathway ID | Pathway Name | p-Value | Adjusted p-Value | Enriched Genes |
|---|---|---|---|---|
| hsa00052 | Galactose metabolism_Homo sapiens_hsa00052 | 7.855E-03 | 1.411E-02 | GAA; AKR1B1 |
| hsa00140 | Steroid hormone biosynthesis_Homo sapiens_hsa00140 | 6.798E-09 | 1.397E-07 | HSD17B1; CYP1A2; HSD17B2; CYP1A1; CYP1B1; CYP3A4; CYP19A1 |
| hsa00232 | Caffeine metabolism_Homo sapiens_hsa00232 | 1.941E-04 | 5.887E-04 | CYP1A2; XDH |
| hsa00350 | Tyrosine metabolism_Homo sapiens_hsa00350 | 1.059E-02 | 1.814E-02 | MAOA; TYR |
| hsa00380 | Tryptophan metabolism_Homo sapiens_hsa00380 | 2.962E-05 | 1.183E-04 | MAOA; CYP1A2; CYP1A1; CYP1B1 |
| hsa00500 | Starch and sucrose metabolism_Homo sapiens_hsa00500 | 2.581E-02 | 3.979E-02 | GAA; PYGL |
| hsa00590 | Arachidonic acid metabolism_Homo sapiens_hsa00590 | 1.096E-08 | 2.028E-07 | CYP2C9; CYP2C8; PLA2G1B; ALOX5; ALOX15; ALOX12; CYP2C19 |
| hsa00591 | Linoleic acid metabolism_Homo sapiens_hsa00591 | 3.917E-11 | 1.811E-09 | CYP2C9; CYP2C8; PLA2G1B; CYP1A2; ALOX15; CYP3A4; CYP2C19 |
| hsa00830 | Retinol metabolism_Homo sapiens_hsa00830 | 1.043E-05 | 4.946E-05 | CYP2C9; CYP2C8; CYP1A2; CYP1A1; CYP3A4 |
| hsa00910 | Nitrogen metabolism_Homo sapiens_hsa00910 | 3.425E-20 | 6.336E-18 | CA12; CA1; CA3; CA2; CA5A; CA4; CA7; CA6; CA9; CA14 |
Showing 1 to 10 of 123 entries
Human Diseases Associated with the Target Proteins of the Plant (Activity≤10μM)
Detailed Information of Human Diseases
| ICD10 Disease Category | Disease Name | ICD10 Code | Linked Targets |
|---|---|---|---|
| A00-B99: Certain infectious and parasitic diseases | Fungal infections | B35-B49 | ALOX5 |
| A00-B99: Certain infectious and parasitic diseases | Infections disease | A00-B99 | MPO |
| A00-B99: Certain infectious and parasitic diseases | Malaria | B54 | CYP2D6 |
| A00-B99: Certain infectious and parasitic diseases | Visceral leishmaniasis | B55.0 | PLA2G1B |
| A00-B99: Certain infectious and parasitic diseases | Leishmaniasis | B55.0 | PLA2G1B |
| A00-B99: Certain infectious and parasitic diseases | Bacterial infections | A00-B99 | CYP2C9 |
| A00-B99: Certain infectious and parasitic diseases | Lymphatic filariasis | B74 | ALOX5 |
| A00-B99: Certain infectious and parasitic diseases | HCV infection | B17.1, B18.2 | MMP3 |
| A00-B99: Certain infectious and parasitic diseases | Pediculus humanus capitis | B85.0 | ACHE |
| A00-B99: Certain infectious and parasitic diseases | Human immunodeficiency virus infection | B20-B26 | FLT3 |
Showing 1 to 10 of 219 entries