Collective Molecular Activities of the Plant: Garcinia Pseudoguttifera
Plant ID: NPO26677
Plant Latin Name: Garcinia Pseudoguttifera
Taxonomy Genus: Garcinia
Taxonomy Family: Clusiaceae
Plant External Links:
NCBI TaxonomyDB:
n.a.
Plant-of-the-World-Online:
n.a.
Country/Region:
ChinaTraditional Medicine System:
TCMChina
Overview of Ingredients
66 All known Ingredients in Total
Unique ingredients have been isolated from this plant.Plant-Ingredients Associations were manually curated from publications or collected from other databases.
44 Ingredients with Acceptable Bioavailablity
Unique ingredients exhibit acceptable human oral bioavailablity, according to the criteria of SwissADME [PMID: 28256516] and HobPre [PMID: 34991690]. The criteria details:SwissADME: six descriptors are used by SwissADME to evaluate the oral bioavailability of a natural product:
☑ LIPO(Lipophility): -0.7 < XLOGP3 < +5.0
☑ SIZE: 150g/mol < MW < 500g/mol
☑ POLAR(Polarity): 20Ų < TPSA < 130Ų
☑ INSOLU(Insolubility): -6 < Log S (ESOL) < 0
☑ INSATU(Insaturation): 0.25 < Fraction Csp3 < 1
☑ FLEX(Flexibility): 0 < Num. rotatable bonds < 9
If 6 descriptors of a natural plant satisfy the above rules, it will be labeled high HOB.
HobPre: A natural plant ingredient with HobPre score >0.5 is labeled high human oral availability (HOB)
26 Ingredients with experimental-derived Activity
Unique ingredients have activity data available.Ingredient Structrual Cards
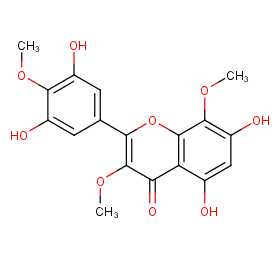
Ingredient ID: NPC99068
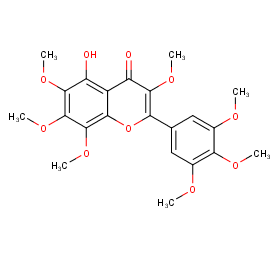
Ingredient ID: NPC85561
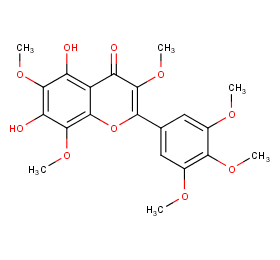
Ingredient ID: NPC7973
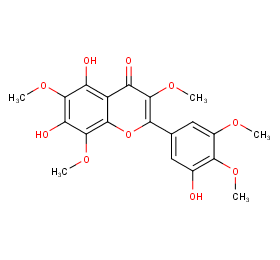
Ingredient ID: NPC78302
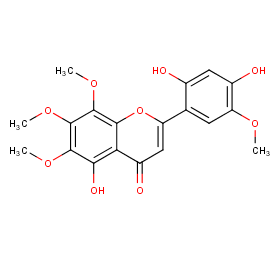
Ingredient ID: NPC7466
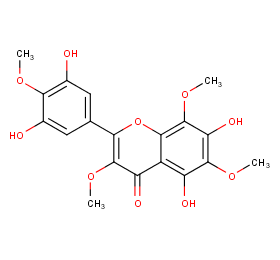
Ingredient ID: NPC74580
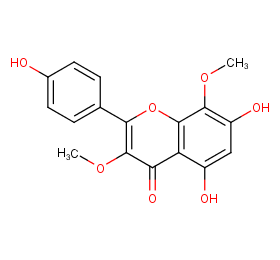
Ingredient ID: NPC70036
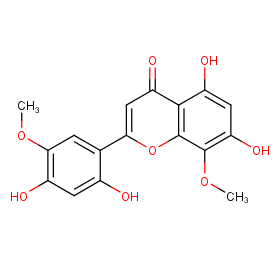
Ingredient ID: NPC67195
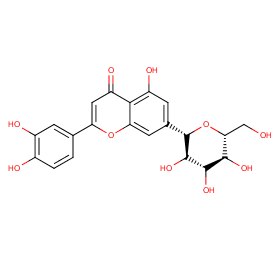
Ingredient ID: NPC65435
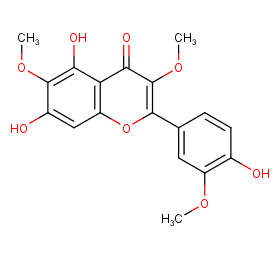
Ingredient ID: NPC55900
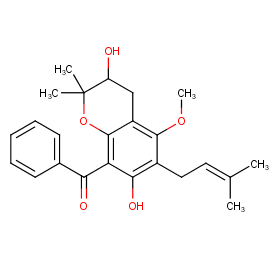
Ingredient ID: NPC51819
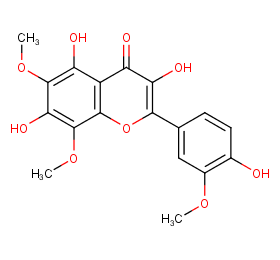
Ingredient ID: NPC49448
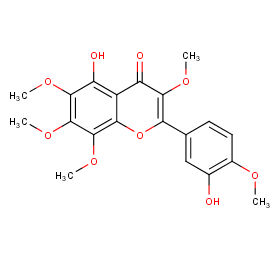
Ingredient ID: NPC4481
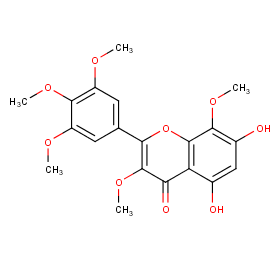
Ingredient ID: NPC43412
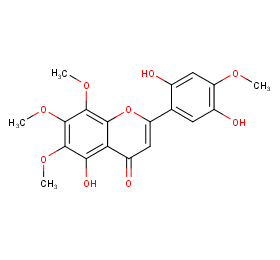
Ingredient ID: NPC312617
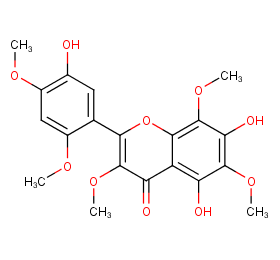
Ingredient ID: NPC310934
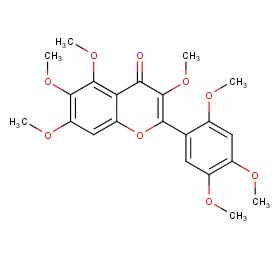
Ingredient ID: NPC299565
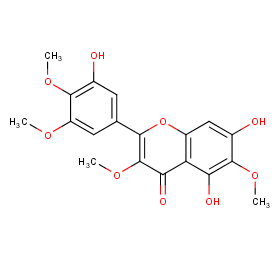
Ingredient ID: NPC29841
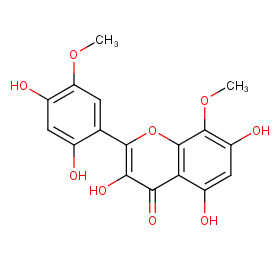
Ingredient ID: NPC293539
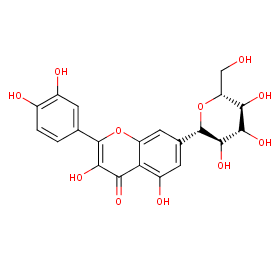
Ingredient ID: NPC292902
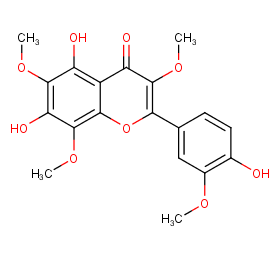
Ingredient ID: NPC288669
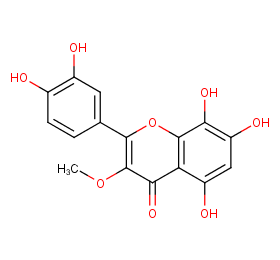
Ingredient ID: NPC287979
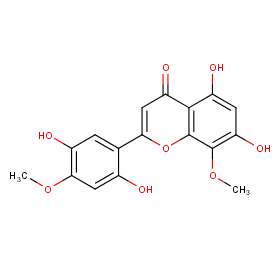
Ingredient ID: NPC287865
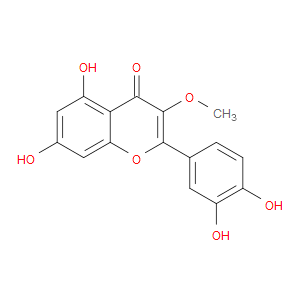
Ingredient ID: NPC286342
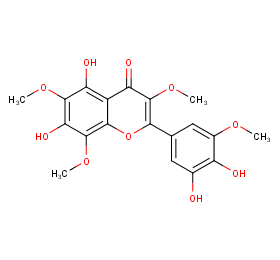
Ingredient ID: NPC286170
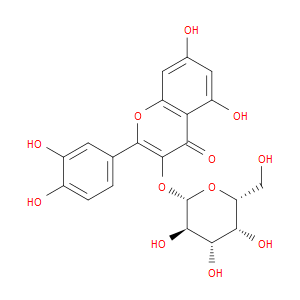
Ingredient ID: NPC281131
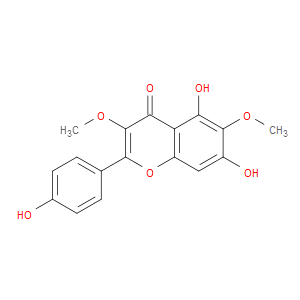
Ingredient ID: NPC266960
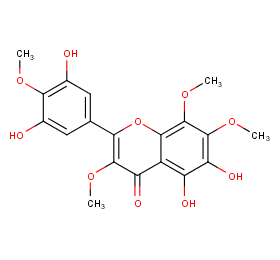
Ingredient ID: NPC26596
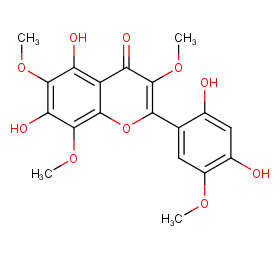
Ingredient ID: NPC26496
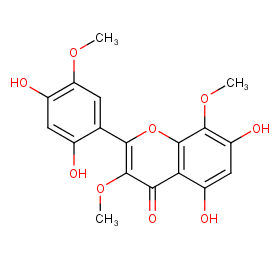
Ingredient ID: NPC262756
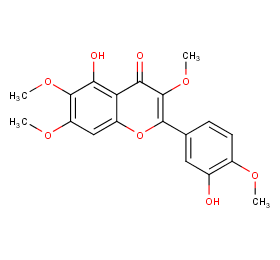
Ingredient ID: NPC25495
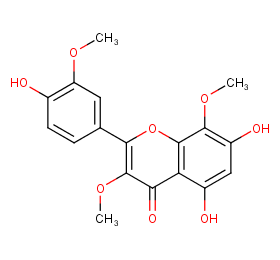
Ingredient ID: NPC253634
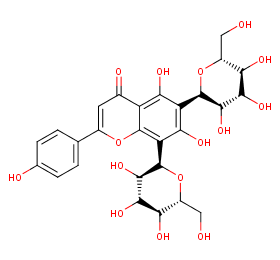
Ingredient ID: NPC238192
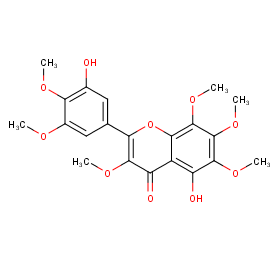
Ingredient ID: NPC235215

Ingredient ID: NPC22472

Ingredient ID: NPC22449
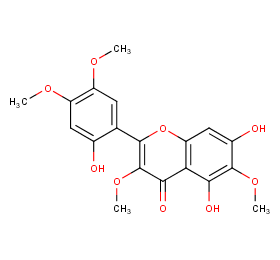
Ingredient ID: NPC222750
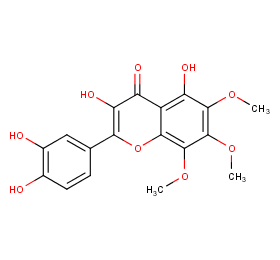
Ingredient ID: NPC221388
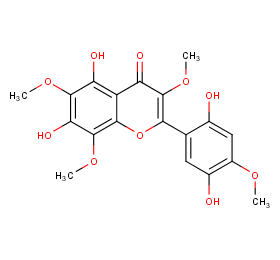
Ingredient ID: NPC219737
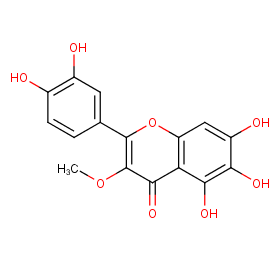
Ingredient ID: NPC214138
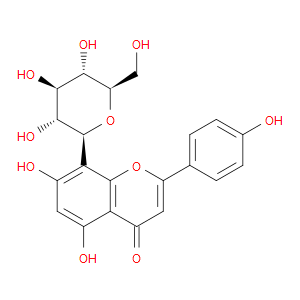
Ingredient ID: NPC209846
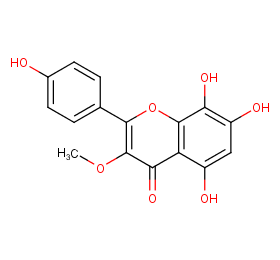
Ingredient ID: NPC208683
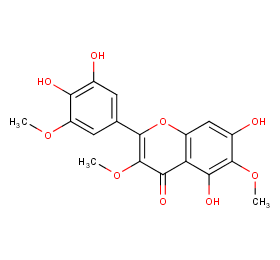
Ingredient ID: NPC207691
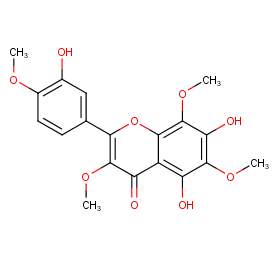
Ingredient ID: NPC204854
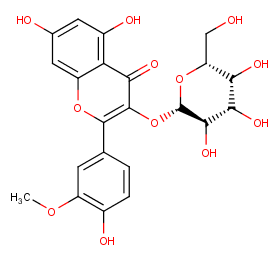
Ingredient ID: NPC203050
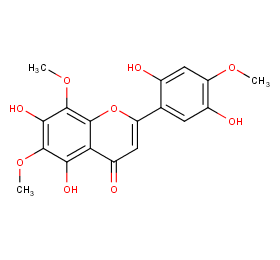
Ingredient ID: NPC197499
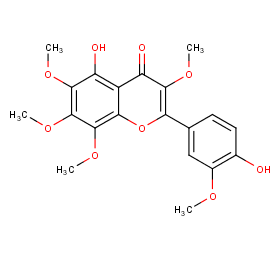
Ingredient ID: NPC19687
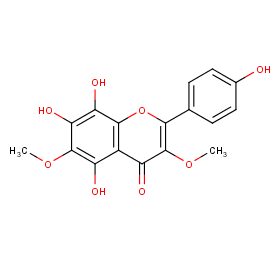
Ingredient ID: NPC195046
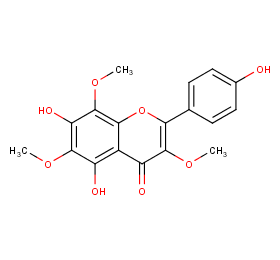
Ingredient ID: NPC191459
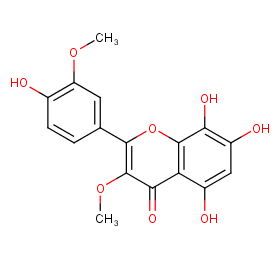
Ingredient ID: NPC190322
Ingredient ID: NPC189958
Ingredient ID: NPC185170
Ingredient ID: NPC176665
Ingredient ID: NPC166778
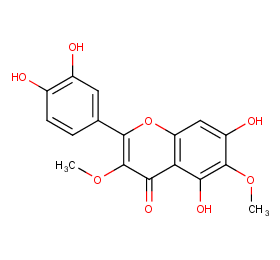
Ingredient ID: NPC163524
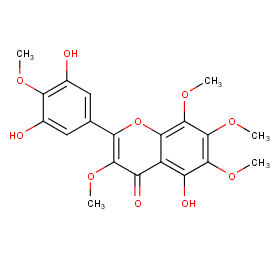
Ingredient ID: NPC142771
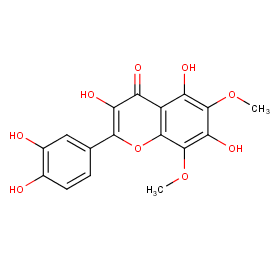
Ingredient ID: NPC137915
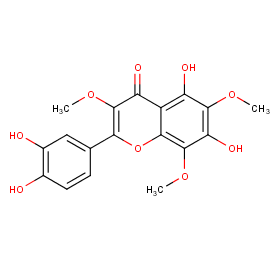
Ingredient ID: NPC130894
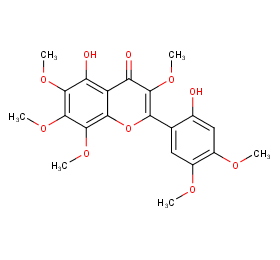
Ingredient ID: NPC123557
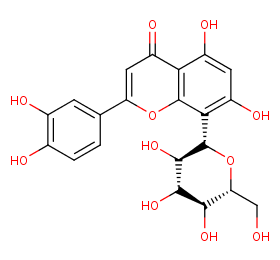
Ingredient ID: NPC117418
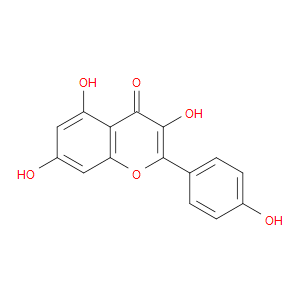
Ingredient ID: NPC116775
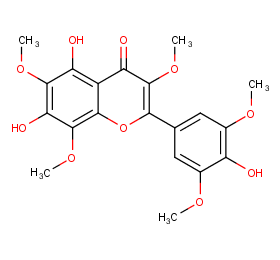
Ingredient ID: NPC1157
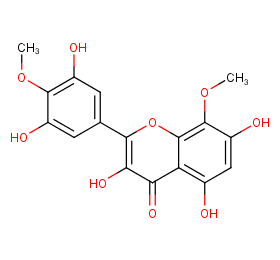
Ingredient ID: NPC115245
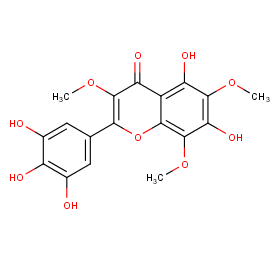
Ingredient ID: NPC114132
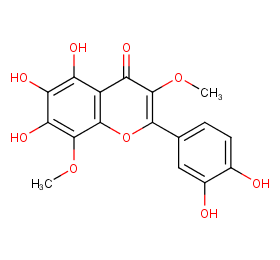
Ingredient ID: NPC10980
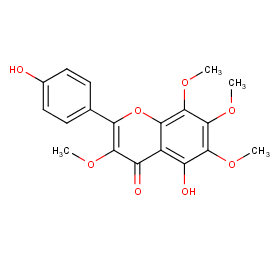
Ingredient ID: NPC105242
Classification of Human Proteins Collectively Targeted by the Plant
Detailed Information of Target Proteins
| Target Type | Protein Class | Gene ID | Protein Name | Uniprot ID | Target ChEMBL ID |
|---|---|---|---|---|---|
| Cytochrome P450 | Cytochrome P450 family 1 | CYP1B1 | Cytochrome P450 1B1 | Q16678 | CHEMBL4878 |
| Cytochrome P450 | Cytochrome P450 family 1 | CYP1A1 | Cytochrome P450 1A1 | P04798 | CHEMBL2231 |
| Cytochrome P450 | Cytochrome P450 family 1 | CYP1A2 | Cytochrome P450 1A2 | P05177 | CHEMBL3356 |
| Therapeutic Target | Enzyme | ALOX12 | Arachidonate 12-lipoxygenase | P18054 | CHEMBL3687 |
| Therapeutic Target | Enzyme | POLB | DNA polymerase beta | P06746 | CHEMBL2392 |
| Therapeutic Target | Enzyme | NOX4 | NADPH oxidase 4 | Q9NPH5 | CHEMBL1250375 |
| Therapeutic Target | Lyase | CA12 | Carbonic anhydrase XII | O43570 | CHEMBL3242 |
| Therapeutic Target | Lyase | CA7 | Carbonic anhydrase VII | P43166 | CHEMBL2326 |
| Therapeutic Target | Oxidoreductase | HSD17B2 | Estradiol 17-beta-dehydrogenase 2 | P37059 | CHEMBL2789 |
| Therapeutic Target | Oxidoreductase | HSD17B10 | Endoplasmic reticulum-associated amyloid beta-peptide-binding protein | Q99714 | CHEMBL4159 |
| Therapeutic Target | Peptide receptor (family A GPCR) | NPSR1 | Neuropeptide S receptor | Q6W5P4 | CHEMBL5162 |
| Therapeutic Target | Protein Kinase | IGF1R | Insulin-like growth factor I receptor | P08069 | CHEMBL1957 |
| Therapeutic Target | Protein Kinase | FLT3 | Tyrosine-protein kinase receptor FLT3 | P36888 | CHEMBL1974 |
| Therapeutic Target | Protein Kinase | PIM1 | Serine/threonine-protein kinase PIM1 | P11309 | CHEMBL2147 |
| Therapeutic Target | Protein Kinase | YES1 | Tyrosine-protein kinase YES | P07947 | CHEMBL2073 |
| Therapeutic Target | Protein Kinase | AURKB | Serine/threonine-protein kinase Aurora-B | Q96GD4 | CHEMBL2185 |
| Therapeutic Target | Protein Kinase | CDK1 | Cyclin-dependent kinase 1 | P06493 | CHEMBL308 |
| Therapeutic Target | Protein Kinase | MET | Hepatocyte growth factor receptor | P08581 | CHEMBL3717 |
| Therapeutic Target | Protein Kinase | KDR | Vascular endothelial growth factor receptor 2 | P35968 | CHEMBL279 |
| Therapeutic Target | Protein Kinase | AXL | Tyrosine-protein kinase receptor UFO | P30530 | CHEMBL4895 |
| Therapeutic Target | Transcription factor | AHR | Aryl hydrocarbon receptor | P35869 | CHEMBL3201 |
Clinical trials associated with plant from natural product (NP) & plant level:
| Clinical trials type | Number of clinical trials | |
|---|---|---|
| 21 | ||
| NCT ID | Title | Condition | Form in clinical use | Associated by plant or compound |
|---|---|---|---|---|
| NCT02226484 | Can Quercetin Increase Claudin-4 and Improve Esophageal Barrier Function in GERD? | gastroesophageal reflux disease | Quercetin (NPC20791) | |
| NCT04853199 | Quercetin In The Treatment Of SARS-COV 2 | COVID-19 | Quercetin (NPC20791) | |
| NCT01348204 | Nasal Potential Studies Utilizing Cystic Fibrosis Transmembrane Regulator (CFTR) Modulators | cystic fibrosis | Quercetin (NPC20791) | |
| NCT01912820 | Effect of Quercetin on Green Tea Polyphenol Uptake in Prostate Tissue From Patients With Prostate Cancer Undergoing Surgery | prostate adenocarcinoma | Quercetin (NPC20791) | |
| NCT04907253 | Quercetin in Coronary Artery By-pass Surgery | coronary artery disease | Quercetin (NPC20791) | |
| NCT01720147 | Quercetin in Children With Fanconi Anemia; a Pilot Study | Fanconi anemia | Quercetin (NPC20791) | |
| NCT01438320 | Q-Trial in Patients With Hepatitis C | chronic hepatitis C virus infection | Quercetin (NPC20791) | |
| NCT05422885 | Safety and Feasibility of Dasatinib and Quercetin in Adults at Risk for Alzheimer's Disease | aging | Quercetin (NPC20791) | |
| NCT01839344 | Effects of Quercetin on Blood Sugar and Blood Vessel Function in Type 2 Diabetes. | type 2 diabetes mellitus | Quercetin (NPC20791) | |
| NCT03943459 | Sirtuin-1 and Advanced Glycation End-products in Postmenopausal Women With Coronary Disease | coronary artery disease | Quercetin (NPC20791) | |
| NCT05456022 | Therapeutic Efficacy of Quercetin Versus Its Encapsulated Nanoparticle on Tongue Squamous Cell Carcinoma Cell Line | mouth neoplasm | Quercetin (NPC20791) | |
| NCT01708278 | Beneficial Effects of Quercetin in Chronic Obstructive Pulmonary Disease (COPD) | chronic obstructive pulmonary disease | Quercetin (NPC20791) | |
| NCT05062486 | A Study to Evaluate the Safety and Efficacy of RQC for AMD | age-related macular degeneration | Quercetin (NPC20791) | |
| NCT05276895 | Effect of Natural Senolytic Agents & NLRP3 Inhibitors on Osteoarthritis | osteoarthritis | Quercetin (NPC20791) | |
| NCT03989271 | Biological Effects of Quercetin in COPD | chronic obstructive pulmonary disease | Quercetin (NPC20791) | |
| NCT01375101 | Therapeutic Effect of Quercetin and the Current Treatment of Erosive and Atrophic Oral Lichen Planus | oral lichen planus | Quercetin (NPC20791) | |
| NCT04851821 | The Effectiveness of Phytotherapy in SARS-COV2(COVID-19) | COVID-19 | Quercetin (NPC20791) | |
| NCT00455416 | Dietary Intervention in Follicular Lymphoma | follicular lymphoma | Quercetin (NPC20791) | |
| NCT04946383 | Safety and Effectivness of Quercetin & Dasatinib on Epigenetic Aging | aging | Quercetin (NPC20791) | |
| NCT02848131 | Senescence in Chronic Kidney Disease | chronic kidney disease | Quercetin (NPC20791) | |
| NCT04733534 | An Open-Label Intervention Trial to Reduce Senescence and Improve Frailty in Adult Survivors of Childhood Cancer | childhood cancer | Quercetin (NPC20791) |
❱❱❱ Associated Human Diseases and Detailed Association Evidence
How do we define the Plant-Targeted Human Disease Association?
Associated human diseases of an individual plant are summurized based on FOUR types of association evidence, these include:
❶ Association by Therapeutic Target: Bioactive protein targets of the plant were defined in "Molecular Targets" section, target-disease associations collected from TTD database were subsequently used to build the associations between the plant and its targeted human diseases.
❷ Association by Disease Gene Reversion: Plant and a specific disease will be associated when >= 1 plant target gene overlaped with disease's DEGs.
❸ Association by Clinical Trials of Plant: Plant and a specific disease will be associated when >= 1 clinical trial (the plant is the intervetion) can be matched in ClinicalTrials.gov database.
❹ Association by Clinical Trials of Plant Ingredients: Plant and a specific disease will be associated when >= 1 clinical trial (the plant ingredient is the intervetion) can be matched in ClinicalTrials.gov database.
Associated Disease of the Plant |
Association Type & Detailed Evidence |
|---|---|
Breast cancerDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2C60-2C6Y |
KDR,CDK1,EGFR,AXL,CA12,SRC,MET,IGF1R,CYP19A1
|
Other specified malignant neoplasms of kidney, except renal pelvisDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2C90.Y |
AURKB,XDH,TYR,CYP19A1,ALOX5,CYP2C9,MET,ALOX5,AURKB
|
Malignant neoplasms of adrenal glandDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2D11 |
CA12,CDK1,DPP4,NOX4,AURKB,CA7,CYP19A1,MET,HSD17B2
|
Lung cancerDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2C25 |
CDK1,MET,IGF1R,EGFR,KDR,AXL,FLT3,ALOX5
|
Squamous cell carcinoma of bronchus or lungDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2C25.2 |
CA12,CDK1,ALOX12,AURKB,XDH,NPSR1,CA7,CYP2C9
|
Malignant neoplasms of thyroid gland, unspecifiedDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2D10.Z |
CA12,DPP4,XDH,CYP1B1,CA7,ALOX5,MET,NOX4
|
Malignant neoplasms of corpus uteri, unspecifiedDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2C76.Z |
CDK1,AURKB,XDH,NPSR1,ALOX15,CYP2C9,HSD17B2,CYP1A1
|
Malignant neoplasms of thymusDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2C27 |
DPP4,HSD17B10,CISD1,FLT3,CYP1B1,ALOX15,CYP2C9,HSD17B2
|
Glioblastoma of brainDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2A00.00 |
CA12,CSNK2A3,DPP4,TUBB6,CYP1B1,KDR,CYP19A1,MET
|
HepatoblastomaDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2C12.01 |
CA12,CDK1,SRC,AURKB,IGF1R,CYP19A1,NOX4
|
Adenocarcinoma of stomachDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2B72.0 |
CDK1,NOX4,AURKB,NPSR1,ALOX15,CYP19A1,MET
|
Adenocarcinoma of bronchus or lungDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2C25.0 |
CA12,CDK1,AURKB,XDH,NPSR1,CA7,CYP2C9
|
Multiple myelomaDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2A83 |
AXL,SRC,KDR,AHR,IGF1R,FLT3
|
Colorectal cancerDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2B91 |
IGF1R,TP53,FLT3,MET,EGFR,KDR
|
Malignant haematopoietic neoplasmDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2B33 |
CDK1,AURKB,TP53,FLT3,SRC,DPP4
|
Acute myeloid leukaemiaDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2A60 |
FLT3,AXL,SRC,TP53,AURKB,PIM1
|
Liver cancerDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2C12 |
IGF1R,KDR,EGFR,CSNK2A1,MET
|
Brain cancerDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2A00 |
EGFR,MET,SRC,TP53,KDR
|
Ovarian cancerDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2C73 |
SRC,MET,FLT3,KDR,TP53
|
Myelodysplastic syndromeDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2A37 |
KDR,AHR,AXL,TP53,EGFR
|
Other specified malignant neoplasms ofcolonDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2B90.Y |
NOX4,TYR,NPSR1,MET,CYP1A1
|
Malignant neoplasms of oesophagusDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2B70 |
CDK1,AURKB,ALOX15,NPSR1,NOX4
|
Renal cell carcinoma, chromophobe typeDisease Category: X.Extension CodesDisease ICD-11 Code: XH6153 |
CA12,AURKB,CYP1A2,MET,CYP1A1
|
Peritoneal cancerDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2C51 |
KDR,TP53,FLT3,SRC
|
Head and neck cancerDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2D42 |
KDR,TP53,MET,EGFR
|
MelanomaDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2C30 |
TYR,MET,KDR,EGFR
|
Stomach cancerDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2B72 |
TP53,EGFR,KDR,MET
|
Pancreatic cancerDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2C10 |
EGFR,SRC,IGF1R,TP53
|
Mature B-cell leukaemiaDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2A82 |
SRC,TP53,AXL,EGFR
|
PsoriasisDisease Category: 14.Diseases of the skinDisease ICD-11 Code: EA90 |
DPP4,FLT3,ALOX5,AHR
|
West Nile virus infectionDisease Category: 01.Certain infectious or parasitic diseasesDisease ICD-11 Code: 1D46 |
DPP4,CYP19A1,MET,AXL
|
Other specified malignant neoplasms of bronchus or lungDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2C25.Y |
CA12,CDK1,AURKB,XDH
|
Urothelial carcinoma of bladderDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2C94.2 |
CYP1A2,CDK1,CYP2C9,AURKB
|
Chikungunya virus diseaseDisease Category: 01.Certain infectious or parasitic diseasesDisease ICD-11 Code: 1D40 |
EGFR,AXL,XDH,CYP19A1
|
Carpal tunnel syndromeDisease Category: 08.Diseases of the nervous systemDisease ICD-11 Code: 8C10.0 |
DPP4,TUBB6,CYP1B1,NOX4
|
Chronic painDisease Category: 21.Symptoms, signs or clinical findings, not elsewhere classifiedDisease ICD-11 Code: MG30 |
KDR,EGFR,FLT3
|
Pleomorphic sarcomaDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2B54 |
KDR,SRC,MET
|
Non-small cell lung cancerDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2C25 |
KDR,MET,EGFR
|
Renal cell carcinomaDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2C90 |
MET,EGFR,KDR
|
Chronic kidney diseaseDisease Category: 16.Diseases of the genitourinary systemDisease ICD-11 Code: GB61 |
IGF1R,XDH
NCT02848131 |
Cardiovascular diseaseDisease Category: 11.Diseases of the circulatory systemDisease ICD-11 Code: BA00-BE2Z |
SRC,MET,KDR
|
Metastatic lymph node neoplasmDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2D60 |
MET,KDR,EGFR
|
Chronic arterial occlusive diseaseDisease Category: 11.Diseases of the circulatory systemDisease ICD-11 Code: BD4Z |
TP53,DPP4,MET
|
Nasopharyngeal cancerDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2B6B |
ALOX5,CDK1,EGFR
|
ThrombocytopeniaDisease Category: 03.Diseases of the blood or blood-forming organsDisease ICD-11 Code: 3B64 |
ALOX5,AHR,KDR
|
Alzheimer diseaseDisease Category: 08.Diseases of the nervous systemDisease ICD-11 Code: 8A20 |
MET,KDR,FLT3
|
Acute diabete complicationDisease Category: 05.Endocrine, nutritional or metabolic diseasesDisease ICD-11 Code: 5A2Y |
KDR,DPP4,AURKB
|
Adenocarcinoma of prostateDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2C82.0 |
ALOX15,NOX4
NCT01912820 |
PheochromocytomaDisease Category: X.Extension CodesDisease ICD-11 Code: XH3854 |
XDH,MET,NOX4
|
Osteoarthritis, unspecifiedDisease Category: 15.Diseases of the musculoskeletal system or connective tissueDisease ICD-11 Code: FA0Z |
CA12,CDK1,HSD17B2
|
LeishmaniasisDisease Category: 01.Certain infectious or parasitic diseasesDisease ICD-11 Code: 1F54 |
DPP4,NOX4,FLT3
|
Malignant neoplasm metastasis in lymph nodes of head, face or neckDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2D60.0 |
AURKB,NPSR1,NOX4
|
Melanoma of skinDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2C30 |
NOX4,IGF1R,ABCG2
|
Superficial ovarian endometriosisDisease Category: 16.Diseases of the genitourinary systemDisease ICD-11 Code: GA10.B4 |
DPP4,TUBB6,SRC
|
Cytomegaloviral diseaseDisease Category: 01.Certain infectious or parasitic diseasesDisease ICD-11 Code: 1D82 |
ALOX15,CYP2C9,CYP1A1
|
Invasive carcinoma of breastDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2C61 |
CA12,CDK1,AURKB
|
Human immunodeficiency virus disease without mention of tuberculosis or malariaDisease Category: 01.Certain infectious or parasitic diseasesDisease ICD-11 Code: 1C62 |
AXL,MET,FLT3
|
Ebola virus diseaseDisease Category: 01.Certain infectious or parasitic diseasesDisease ICD-11 Code: 1D60.01 |
AXL,MET,HSD17B2
|
Mesothelioma of pleuraDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2C26.0 |
RECQL,EGFR,XDH,FLT3,AHR,MET,CA12,CDK1,AURKB,TYR,PIM1,KDR,CA7,HSD17B2,DPP4,SRC,CSNK2A2,ALOX15,ALOX5,AXL
|
Endometrial cancerDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2C76 |
SRC,TP53
|
Mature B-cell lymphomaDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2A85 |
FLT3,CDK1
|
InfluenzaDisease Category: 01.Certain infectious or parasitic diseasesDisease ICD-11 Code: 1E30-1E32 |
XDH,ALOX5
|
Myeloproliferative neoplasmDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2A20 |
SRC,FLT3
|
BCR-ABL1-negative chronic myeloid leukaemiaDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2A41 |
SRC,FLT3
|
Pituitary gland disorderDisease Category: 05.Endocrine, nutritional or metabolic diseasesDisease ICD-11 Code: 5A60-5A61 |
IGF1R,ALOX5
|
Unspecific body region injuryDisease Category: 22.Injury, poisoning or certain other consequences of external causesDisease ICD-11 Code: ND56 |
MET,EGFR
|
Postoperative inflammationDisease Category: 01.Certain infectious or parasitic diseasesDisease ICD-11 Code: 1A00-CA43 |
ALOX5,SRC
|
Type 2 diabetes mellitusDisease Category: 05.Endocrine, nutritional or metabolic diseasesDisease ICD-11 Code: 5A11 |
DPP4
NCT01839344 |
Cervical cancerDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2C77 |
SRC,TP53
|
Pain, unspecifiedDisease Category: 21.Symptoms, signs or clinical findings, not elsewhere classifiedDisease ICD-11 Code: MG3Z |
ALOX5,IGF1R
|
Rheumatoid arthritisDisease Category: 15.Diseases of the musculoskeletal system or connective tissueDisease ICD-11 Code: FA20 |
ABCG2,ALOX5
|
Idiopathic interstitial pneumonitisDisease Category: 12.Diseases of the respiratory systemDisease ICD-11 Code: CB03 |
ALOX5,FLT3
|
Bacterial infectionDisease Category: 01.Certain infectious or parasitic diseasesDisease ICD-11 Code: 1A00-1C4Z |
CA12,CYP2C9
|
IschemiaDisease Category: 08.Diseases of the nervous systemDisease ICD-11 Code: 8B10-8B11 |
EGFR,SRC
|
Motor neuron diseaseDisease Category: 08.Diseases of the nervous systemDisease ICD-11 Code: 8B60 |
IGF1R,SRC
|
Gastrointestinal stromal tumourDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2B5B |
SRC,KDR
|
Fallopian tube cancerDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2C74 |
TP53,SRC
|
NADisease Category: NADisease ICD-11 Code: NA |
MET,NOX4
|
Atopic eczemaDisease Category: 14.Diseases of the skinDisease ICD-11 Code: EA80 |
AHR,ALOX5
|
Malignant digestive organ neoplasmDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2C11 |
KDR,SRC
|
Thyroid cancerDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2D10 |
MET,KDR
|
coronary artery diseaseDisease Category: 11.Diseases of the circulatory systemDisease ICD-11 Code: BA8Y-BA8Z |
NCT03943459,NCT04907253
|
COVID-19Disease Category: 01.Certain infectious or parasitic diseasesDisease ICD-11 Code: 1D6Y |
NCT04851821,NCT04853199
|
Ageing associated decline in intrinsic capacityDisease Category: 21.Symptoms, signs or clinical findings, not elsewhere classifiedDisease ICD-11 Code: MG2A |
NCT04946383,NCT05422885
|
Chronic obstructive pulmonary diseaseDisease Category: 12.Diseases of the respiratory systemDisease ICD-11 Code: CA22 |
NCT01708278,NCT03989271
|
Coeliac diseaseDisease Category: 13.Diseases of the digestive systemDisease ICD-11 Code: DA95 |
CDK1,CYP1A1
|
Myelodysplastic syndromes, unspecifiedDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2A3Z |
PIM1,CYP2C9
|
Dengue fever, unspecifiedDisease Category: 01.Certain infectious or parasitic diseasesDisease ICD-11 Code: 1D2Z |
ABCG2,AURKB
|
Systemic lupus erythematosusDisease Category: 04.Diseases of the immune systemDisease ICD-11 Code: 4A40.0 |
YES1,CYP1A1
|
Heart failure, unspecifiedDisease Category: 11.Diseases of the circulatory systemDisease ICD-11 Code: BD1Z |
ABCG2,CYP1B1
|
Gliomas of brainDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2A00.0 |
AHR,NOX4
|
Solid tumour/cancerDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2A00-2F9Z |
FLT3,CSNK2A1,XDH,AURKB,AHR,AXL,PIM1,MET,SRC,POLB,ALOX5,CDK1,IGF1R,KDR,YES1,TP53,EGFR,CA12
|
Adenocarcinoma of pancreasDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2C10.0 |
DPP4,RECQL,POLB,AURKB,CISD1,AXL,ABCG2,XDH,NPSR1,TUBB6,PIM1,CYP1B1,CA7,ALOX15,ALOX5,CYP2C9,MET,HSD17B2
|
Diffuse large B-cell lymphomasDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2A81 |
RECQL,HSD17B10,EGFR,AHR,MET,CA12,CDK1,TP53,AURKB,KDR,SRC,CISD1,CSNK2A2,AXL,TUBB6,NOX4
|
Carcinosarcoma of uterusDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2C76.43 |
DPP4,TP53,POLB,HSD17B10,AURKB,CISD1,XDH,NPSR1,YES1,CA7,ALOX15,CYP19A1,MET,HSD17B2
|
Serous cystadenoma,borderline malignancy of ovaryDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2C73.4 |
CA12,CSNK2A3,NOX4,ALOX12,TP53,CISD1,ALOX15,XDH,YES1,CYP1A2,AHR,ALOX5,MET,HSD17B2
|
Germ cell tumour of testisDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2C80.2 |
CA12,CDK1,DPP4,ALOX12,NOX4,ABCG2,XDH,NPSR1,KDR,AHR,ALOX5,CYP2C9,MET,HSD17B2
|
Malignant neoplasms of biliary tract, distal bile ductDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2C15 |
CSNK2A3,CDK1,ALOX12,TP53,SRC,AURKB,IGF1R,TUBB6,ALOX5,NOX4
|
Hepatocellular carcinoma of liverDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2C12.02 |
CA12,CDK1,AURKB,CYP19A1,NOX4,CA12,CDK1,SRC,AURKB,CYP19A1
|
Diabetes mellitus, type unspecifiedDisease Category: 05.Endocrine, nutritional or metabolic diseasesDisease ICD-11 Code: 5A14 |
DPP4
|
Chronic myelomonocytic leukaemiaDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2A40 |
FLT3
|
Esophageal cancerDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2B70 |
TP53
|
Intracerebral haemorrhageDisease Category: 08.Diseases of the nervous systemDisease ICD-11 Code: 8B00 |
EGFR
|
Metastatic digestive system neoplasmDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2D8Y |
SRC
|
Human immunodeficiency virus diseaseDisease Category: 01.Certain infectious or parasitic diseasesDisease ICD-11 Code: 1C60-1C62 |
FLT3
|
EncephalopathyDisease Category: 08.Diseases of the nervous systemDisease ICD-11 Code: 8E47 |
XDH
|
Seborrhoeic dermatitisDisease Category: 14.Diseases of the skinDisease ICD-11 Code: EA81 |
CA12
|
Biliary tract cancerDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2C17 |
MET
|
Inborn purine/pyrimidine/nucleotide metabolism errorDisease Category: 05.Endocrine, nutritional or metabolic diseasesDisease ICD-11 Code: 5C55 |
XDH
|
Mineral deficiencyDisease Category: 05.Endocrine, nutritional or metabolic diseasesDisease ICD-11 Code: 5B5K |
XDH
|
Malignant mesenchymal neoplasmDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2B5D-2B5Y |
EGFR
|
Fungal infectionDisease Category: 01.Certain infectious or parasitic diseasesDisease ICD-11 Code: 1F29-1F2F |
ALOX5
|
Diabetic foot ulcerDisease Category: 11.Diseases of the circulatory systemDisease ICD-11 Code: BD54 |
EGFR
|
FilariasisDisease Category: 01.Certain infectious or parasitic diseasesDisease ICD-11 Code: 1F66 |
ALOX5
|
Liver diseaseDisease Category: 13.Diseases of the digestive systemDisease ICD-11 Code: DB90-DB9Z |
MET
|
LiposarcomaDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2B59 |
TP53
|
Bone/articular cartilage neoplasmDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2F7B |
FLT3
|
GoutDisease Category: 15.Diseases of the musculoskeletal system or connective tissueDisease ICD-11 Code: FA25 |
XDH
|
MastocytosisDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2A21 |
FLT3
|
GlaucomaDisease Category: 09.Diseases of the visual systemDisease ICD-11 Code: 9C61 |
MET
|
AsthmaDisease Category: 12.Diseases of the respiratory systemDisease ICD-11 Code: CA23 |
ALOX5
|
Autism spectrum disorderDisease Category: 06.Mental, behavioural or neurodevelopmental disordersDisease ICD-11 Code: 6A02 |
DPP4
|
Coronavirus infectionDisease Category: 01.Certain infectious or parasitic diseasesDisease ICD-11 Code: 1D92 |
CSNK2A1
|
RetinopathyDisease Category: 09.Diseases of the visual systemDisease ICD-11 Code: 9B71 |
IGF1R
|
Multiple structural anomalies syndromeDisease Category: 20.Developmental anomaliesDisease ICD-11 Code: LD2F |
IGF1R
|
Extraocular muscle disorderDisease Category: 09.Diseases of the visual systemDisease ICD-11 Code: 9C82 |
IGF1R
|
LeiomyosarcomaDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2B58 |
SRC
|
Colon cancerDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2B90 |
EGFR
|
Epidermal dysplasiasDisease Category: 14.Diseases of the skinDisease ICD-11 Code: EK90 |
SRC
|
Testicular cancerDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2C80 |
EGFR
|
Lip/oral cavity/pharynx neoplasmDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2B6E |
TP53
|
Ovarian dysfunctionDisease Category: 05.Endocrine, nutritional or metabolic diseasesDisease ICD-11 Code: 5A80 |
TP53
|
Acquired hypomelanotic disorderDisease Category: 14.Diseases of the skinDisease ICD-11 Code: ED63 |
TYR
|
Ureteral cancerDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2C92 |
AHR
|
Carcinoid syndromeDisease Category: 05.Endocrine, nutritional or metabolic diseasesDisease ICD-11 Code: 5B10 |
ALOX5
|
Mature T-cell lymphomaDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2A90 |
EGFR
|
Hepatitis virus infectionDisease Category: 01.Certain infectious or parasitic diseasesDisease ICD-11 Code: 1E50-1E51 |
EGFR
|
Spinal cord/cranial nerve neoplasmDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2A02 |
EGFR
|
Urethral cancerDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2C93 |
EGFR
|
Metastatic liver/bile duct neoplasmDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2D80 |
TP53
|
Angina pectorisDisease Category: 11.Diseases of the circulatory systemDisease ICD-11 Code: BA40 |
EGFR
|
Synovial sarcomaDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2B5A |
SRC
|
Inflammatory arthropathyDisease Category: 15.Diseases of the musculoskeletal system or connective tissueDisease ICD-11 Code: FA2Z |
FLT3
|
Myocardial infarctionDisease Category: 11.Diseases of the circulatory systemDisease ICD-11 Code: BA41-BA43 |
DPP4
|
Multiple sclerosisDisease Category: 08.Diseases of the nervous systemDisease ICD-11 Code: 8A40 |
SRC
|
Metastatic malignant neoplasmDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2D50-2E09 |
AXL
|
Phakomatoses/hamartoneoplastic syndromeDisease Category: 20.Developmental anomaliesDisease ICD-11 Code: LD2D |
FLT3
|
Kidney failureDisease Category: 16.Diseases of the genitourinary systemDisease ICD-11 Code: GB60-GB6Z |
TP53
|
ParkinsonismDisease Category: 08.Diseases of the nervous systemDisease ICD-11 Code: 8A00 |
SRC
|
Metastatic biliary tract neoplasmsDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2C14-2C17 |
EGFR
|
Irritable bowel syndromeDisease Category: 13.Diseases of the digestive systemDisease ICD-11 Code: DD91 |
ABCG2
|
Skin postprocedural disorderDisease Category: 14.Diseases of the skinDisease ICD-11 Code: EL8Y |
EGFR
|
Cushing syndromeDisease Category: 05.Endocrine, nutritional or metabolic diseasesDisease ICD-11 Code: 5A70 |
CYP19A1
|
Diffuse large B-cell lymphomaDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2A81 |
FLT3
|
EndometriosisDisease Category: 16.Diseases of the genitourinary systemDisease ICD-11 Code: GA10 |
CYP19A1
|
Heart failureDisease Category: 11.Diseases of the circulatory systemDisease ICD-11 Code: BD10-BD1Z |
XDH
|
Digestive organ benign neoplasmDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2E92 |
KDR
|
Transplant rejectionDisease Category: 22.Injury, poisoning or certain other consequences of external causesDisease ICD-11 Code: NE84 |
TP53
|
Ischemic heart diseaseDisease Category: 11.Diseases of the circulatory systemDisease ICD-11 Code: BA40-BA6Z |
MET
|
ThyrotoxicosisDisease Category: 05.Endocrine, nutritional or metabolic diseasesDisease ICD-11 Code: 5A02 |
IGF1R
|
Hypopharyngeal cancerDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2B6D |
EGFR
|
OsteosarcomaDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2B51 |
SRC
|
OesophagitisDisease Category: 13.Diseases of the digestive systemDisease ICD-11 Code: DA24 |
CYP19A1
|
Prostate cancerDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2C82 |
AURKB
|
Bladder cancerDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2C94 |
TP53
|
Acquired hypermelanosisDisease Category: 14.Diseases of the skinDisease ICD-11 Code: ED60 |
TYR
|
Metabolic disorderDisease Category: 05.Endocrine, nutritional or metabolic diseasesDisease ICD-11 Code: 5C50-5D2Z |
DPP4
|
Transplanted organ/tissueDisease Category: 24.Factors influencing health status or contact with health servicesDisease ICD-11 Code: QB63 |
FLT3
|
NeuropathyDisease Category: 08.Diseases of the nervous systemDisease ICD-11 Code: 8C0Z |
CYP19A1
|
Unspecified viral infection of unspecified siteDisease Category: 01.Certain infectious or parasitic diseasesDisease ICD-11 Code: 1D9Z |
FLT3
|
Cystic fibrosisDisease Category: 12.Diseases of the respiratory systemDisease ICD-11 Code: CA25 |
NCT01348204
|
OsteoarthritisDisease Category: 15.Diseases of the musculoskeletal system or connective tissueDisease ICD-11 Code: FA00-FA05 |
NCT05276895
|
Lichen planusDisease Category: 14.Diseases of the skinDisease ICD-11 Code: EA91 |
NCT01375101
|
Chronic hepatitis CDisease Category: 01.Certain infectious or parasitic diseasesDisease ICD-11 Code: 1E51.1 |
NCT01438320
|
Unspecified malignant neoplasms of unspecified sitesDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2D4Z |
NCT04733534
|
Malignant neoplasms of other or unspecified parts of mouthDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2B66 |
NCT05456022
|
Gastro-oesophageal reflux diseaseDisease Category: 13.Diseases of the digestive systemDisease ICD-11 Code: DA22 |
NCT02226484
|
Congenital aplastic anaemiaDisease Category: 03.Diseases of the blood or blood-forming organsDisease ICD-11 Code: 3A70.0 |
NCT01720147
|
Follicular lymphomaDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2A80 |
NCT00455416
|
Macular degenerationDisease Category: 09.Diseases of the visual systemDisease ICD-11 Code: 9B75 |
NCT05062486
|
Inflammatory bowel diseases, unspecifiedDisease Category: 13.Diseases of the digestive systemDisease ICD-11 Code: DD7Z |
NOX4
|
Zika virus diseaseDisease Category: 01.Certain infectious or parasitic diseasesDisease ICD-11 Code: 1D48 |
AXL
|
Atrial fibrillationDisease Category: 11.Diseases of the circulatory systemDisease ICD-11 Code: BC81.3 |
CA7
|
Myotonic dystrophyDisease Category: 08.Diseases of the nervous systemDisease ICD-11 Code: 8C71.0 |
CYP1A1
|
Tetralogy of FallotDisease Category: 20.Developmental anomaliesDisease ICD-11 Code: LA88.2 |
ABCG2
|
Pneumonia due to PseudomonasaeruginosaDisease Category: 12.Diseases of the respiratory systemDisease ICD-11 Code: CA40.05 |
DPP4
|
Systemic sclerosisDisease Category: 04.Diseases of the immune systemDisease ICD-11 Code: 4A42 |
NOX4
|
Chronic rhinosinusitisDisease Category: 12.Diseases of the respiratory systemDisease ICD-11 Code: CA0A |
CYP1A1
|
Dilated cardiomyopathyDisease Category: 11.Diseases of the circulatory systemDisease ICD-11 Code: BC43.0 |
ABCG2
|
Lyme borreliosisDisease Category: 01.Certain infectious or parasitic diseasesDisease ICD-11 Code: 1C1G |
MET
|

