Computer Aided Drug Design:
-
Brief introduction of methods.
Mechanism of drug action:

Drug Design Objective:
-
Finding a chemical compound that can
fit to a specific cavity on a protein target both geometrically and chemically.
-
After passing the animal tests and
human clinical trials, this compound becomes a drug available to patients.
Traditional Drug Design Methods:
Random screening of chemicals
found in nature or synthesized in labs.
-
Long design cycle: 7-12 years.
-
High cost: $350 million USD per marketed drug.
Drug Discovery Today 2, 72-78 (1997)
Too slow and costly to meet demand.
Experimental strategies for improving
screening cycle:
-
Smart screening:
High-throughput robotic screening
-
Diversity of chemical compounds:
Nature 384 Suppl., 2-7 (1996)
These methods are yet to show promise.
High cost.
Computer-aided drug design:
-
Protein target 3D structure unknown:
Method:
Quantitative structure activity
relationship (QSAR)
The purpose:
Derive a function that links biological activity of a
group of related chemical compounds with parameters that describe a structural
feature of these molecule.
This feature also reflects the property of binding cavity
on protein target.
The derived function is used as a guide to select best
candidate for drug design.
Biological activity:
The effect of a chemical compound on a biological system
due to its binding to its protein target.
QSAR:

-
Protein target 3D structure known:
Method:
Ligand-protein docking:
Description of the method:
(1) Real ligand-protein binding process


(2) Computer simulation of this ligand-protein binding
process
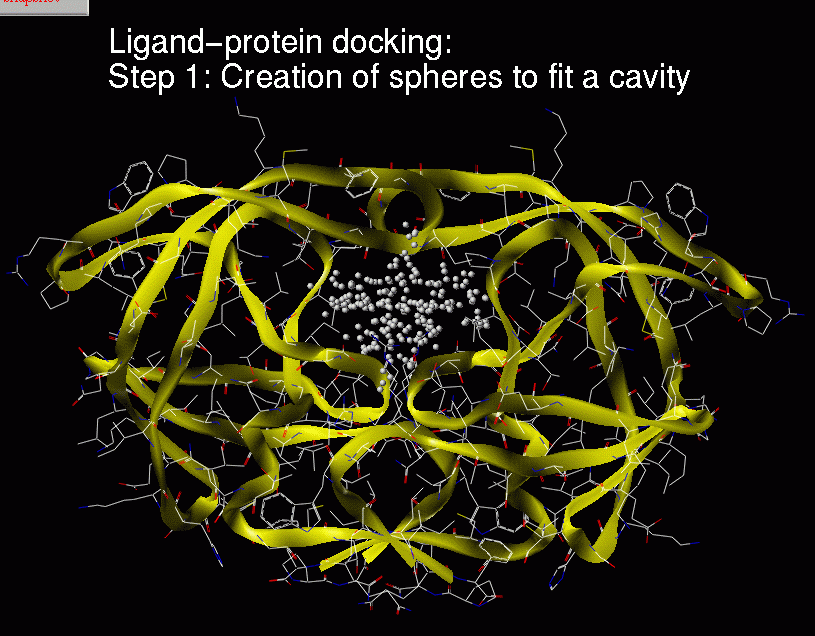
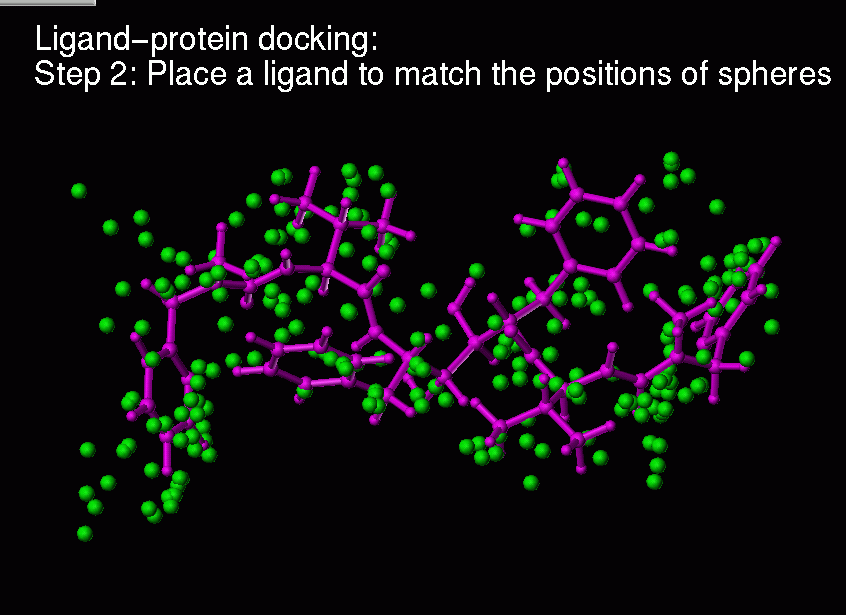
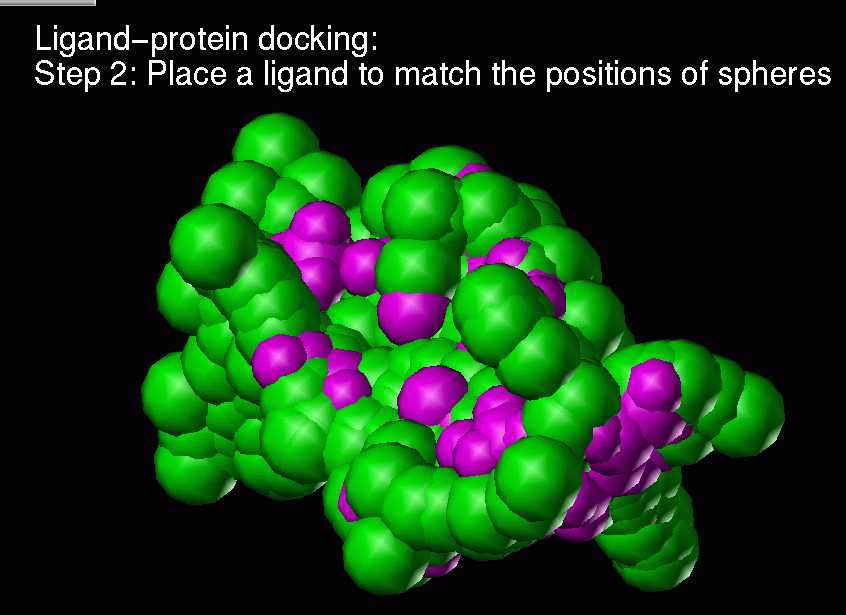
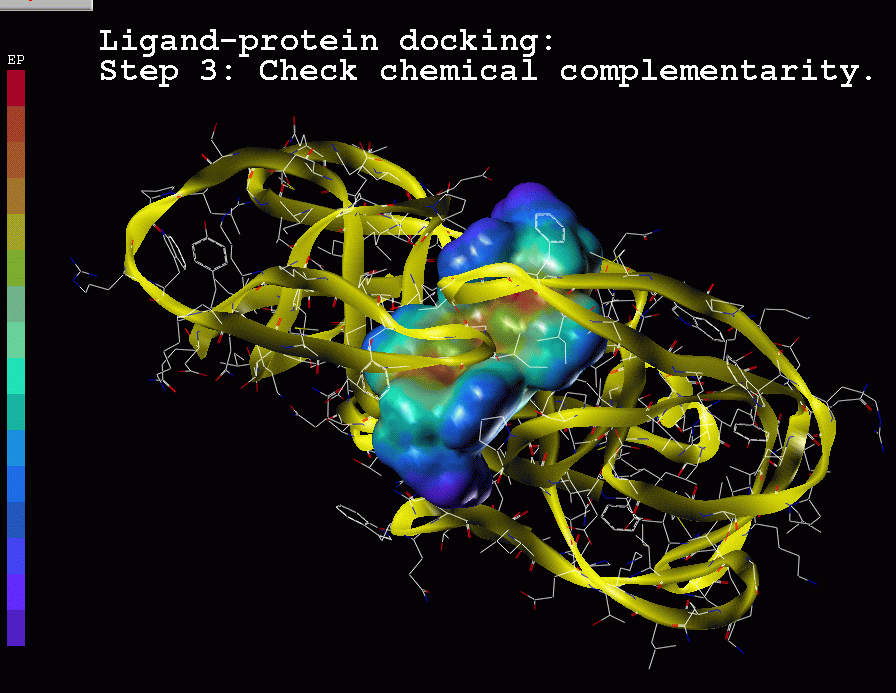
Chemical complementarity is evaluated
based on potential
energy description.
Homework






