 ABOUT NPASS
ABOUT NPASS
Modern drug discovery and today's pharmacopeia is largely benefited from nature. More than 50% of approved drugs are natural products or natural product derivatives. It is estimated that there are about a million natural products have been isolated and many of them have been subjected to experimental assays to evaluate quantitative biological activities. However, there is a lack of an integrated datahouse to assemble these valuable data from individual literatures and provide to research community. With the motivation of addressing this gap, the Natural Product Activity and Species Source Database (NPASS) is developed to provide a freely accessible database integrating detailed information of species sources and biological activities of natural products.
The NPASS database in the current version (V1.0) provides 30,926 unique natural products isolated from 25,041 source organisms and together with 446,552 activity records on 5,863 targets. In addition to isolated NPs, many organisms (whole organisms or certain parts) have been used as medicinal materials in Traditional Chinese Medicine (TCM) and other traditional medicines for a long history. NPASS records also cover chemical ingredients of TCM herbs and their biological activities. For example, at the activity cutoff of <= 10uM, NPASS includes 719 TCM ingredient chemicals against 464 human protein targets (1936 NP-Target pairs). The NPASS will be regularly updated to include more natural products which have both species source and quantitative activity data available from recent publications.
 DATA SOURCES
DATA SOURCES
I. Species source of natural products
Species source information is mainly from manually inspection of publications. Besides, we surveyed existing natural product-related databases to find species source annotations.
♦ Manually annotated from publications
Multiple keywords/keywords conbinations are used to search literatures that may revelant to isolation, total synthesis, activity evaluation of NPs through PubMed. These keywords include natural product, NP, nature, marine, plant, microbe, microbial, bacterium, bacteria, bacterial, fungus, fungi, fungal, species, traditional medicine, medicinal, indigenous, folk, herb, herbal, herbalism, Chinese medicine, TCM, Ayurveda, activity, active, bioactive, potent, potency, IC50, Ki, EC50, GI50, and MIC. Searched publications are subjected to first-step manually check the title to confirm if the literature is really revelant. Then, full articles of these relavent publications are downloaded for manually checking the species source (including if the NP is novel structure claimed by authors, species collection location and time, species part used for isolation, etc.) of corresponding natural products.
♦ Collected from existing databases
Few existing databases include a part of species source information of natural products. These databases includes: TCM-ID, TCMID, TCM@TaiWan, TCMSP, UNPD, TM-MC, StreptomeDB, TTD, TarNet, ChEBI, and HerDing. Therefore, NP names/structures are searched against these databases to extract species source information.
II. Biological activities of natural products
Quantitative activity data of NPs against specific targets (including: target information, activity type and values, compound dose etc.) are curated from literatures searched described in previous paragraph. Collected activity types includes inhibition concentration/dose like IC50/IC90/ID50, activity concentrations like AC/AC40/AC50/Potency, microbial inhibitory or lethal concentrations like MIC/MFC/MBC/FC, growth inhibitory concentrations like GI/GI50/TGI, percentage inhibition at fixed concentrations like inhibition rate, efective contrations/doses like EC50/EC90/ED50/ED90, equilibrium inhibition constant Ki, lethal concentrations/doses like LC/LC50/LC90/LD50/LD90, inhibition zone IZ, equilibrium binding constant Kd, ratio IC50/ratio EC50/ratio/Ki, cytotoxic concentrations like CC25/CC50/CC90/CC100, and toxic concentration/dose like TC50/TD50. About 56% of activity values are stored as the unit of nM, other units include ug/ml, mg/kg, %, mm, and so on.
 NATURAL PRODUCT PROFILE
NATURAL PRODUCT PROFILE
I. Natural products
♦ Natural products chemical representation
Common name, synonyms, IUPAC Name, Standard InCHI, Standard InCHI Key, Canonical SMILES, and MOL file.
♦ Natural products physical & chemical properties
Molecular formula, molecular weight, AlogP, # hydrogen bond donor, # hydrogen bond acceptor, polar surface area, # rotatable bond, # aromatic rings, # heavy atoms.
II. Clinical/approved drugs
Clinical trial and approved drugs are collected from TTD (Therapeutical Target Database), Drug Bank, and ChEMBL database.
III. Similarity between molecules
Structure similarity between molecules is defined by Tanimoto coefficient (Tc). Tc is calculated by using PubChem 881-bit substructure fingerprints according to below equation:
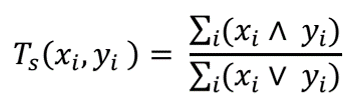
Where 'X' and 'X' are fingerprints of two molecules, and 'xi' and 'yi' are the ith bits in each fingerprint. "∧" and "∨" represent the bitwise "and" and bitwise "or", respectively. Ts(xi,yi) is the value of Tanimoto coefficient which is equal to the total number of common substructure features divided by the total number of unique substructures existing in both molecules.
Tc lies between [0,1] where '1' represents the highest similarity between molecule 'X' and 'Y'.
Tc scores between NPs are pre-calculated and stored in NPASS database. While Tc score between user-query-molecule and NPs is calculated realtime by using functions of chemfp toolkit.
 SOURCE ORGANISM PROFILE
SOURCE ORGANISM PROFILE
I. Organism taxonomy information
All organism names extracted from orginal files are firstly matched to scientific names from NCBI taxonomy database, then unmatched names are matched to synonyms from NCBI Taxonomy Database and transformed to scientific name when matched. Finally, those organism names that can not match to any scientific names or synonyms are kept in orginal format. After matching to NCBI TaxonomyDB, taxonomy IDs are recorded to generate external links of organisms.
II. Organism external links
About 60% and 93% source species can be matched to NCBI Taxonomy database at species level and genus level, respectively. For these species, Taxonomy IDs are annotated so that users can review taxonomic details from NCBI Taxonomy database. For the remaining about 7% species which can not match to any entries in NCBI Taxonomy database, we will further check the accuracy of species name and annotate taxonomic information from original literatures. Apart from NCBI Taxonomy database, species organisms are also links to other databases such as World Register of Marine Species (WoRMS) when data available.
 TARGET PROFILE
TARGET PROFILE
Targets are classified into several categories according to classification of ChEMBL database, including 'Individual protein', 'Protein family', 'Protein complex', 'Protein-protein interaction', 'Cell line', 'Organism', and so on. Targets are corsslinked to TTD, Uniprot, ChEMBL, IUPHAR/BPS when possible.
