Collective Molecular Activities of Useful Plants
Knowledge of the collective activities of individual plants together the derived clinical effects and targeted disease associations is useful for plant-based biomedical research and development. Especially, understanding of the collective activities of individual medicinal plants may aid the development of multi-component therapeutics for complex diseases. These efforts can be benefit from extending collective activities of individual plants to more complex systems such as transcriptome of patients. For example, collective reversion of disease-related transcriptome changes has been demonstrated to be significantly relevant to therapeutic effects of drugs [Ref: Nature communications 8.1 (2017): 16022.].
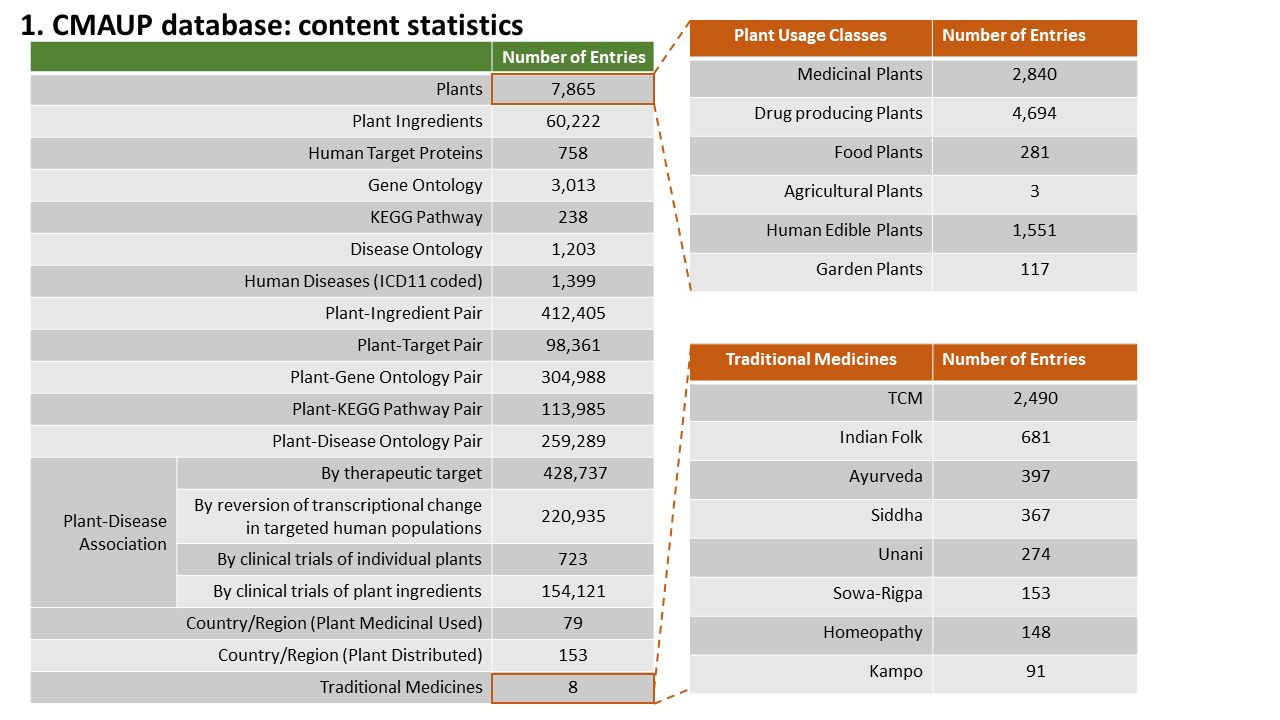
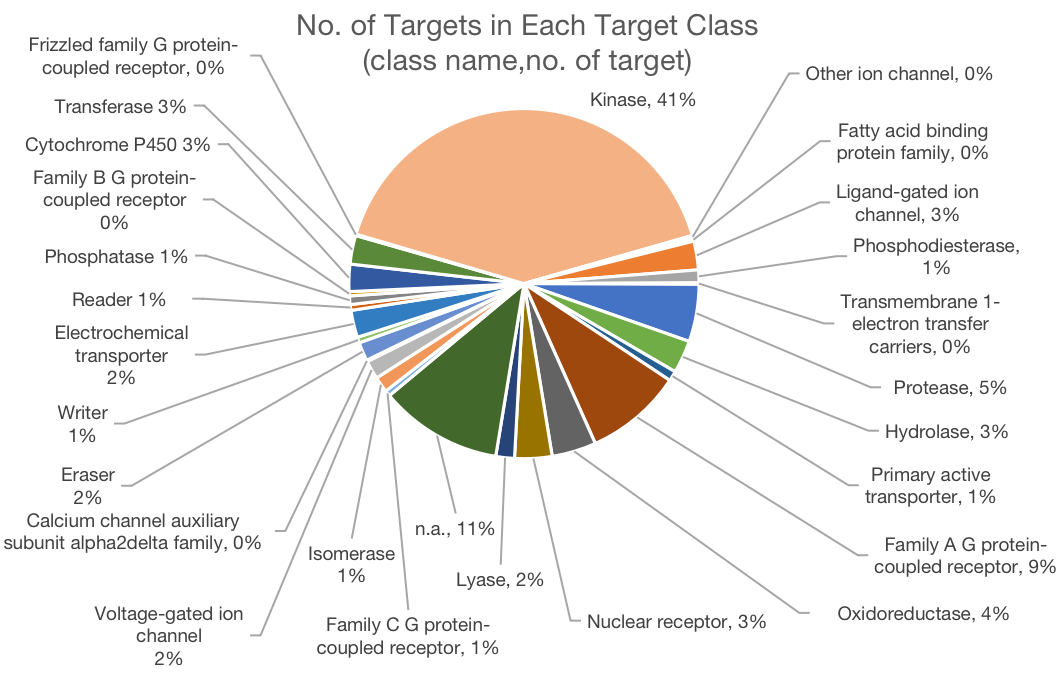
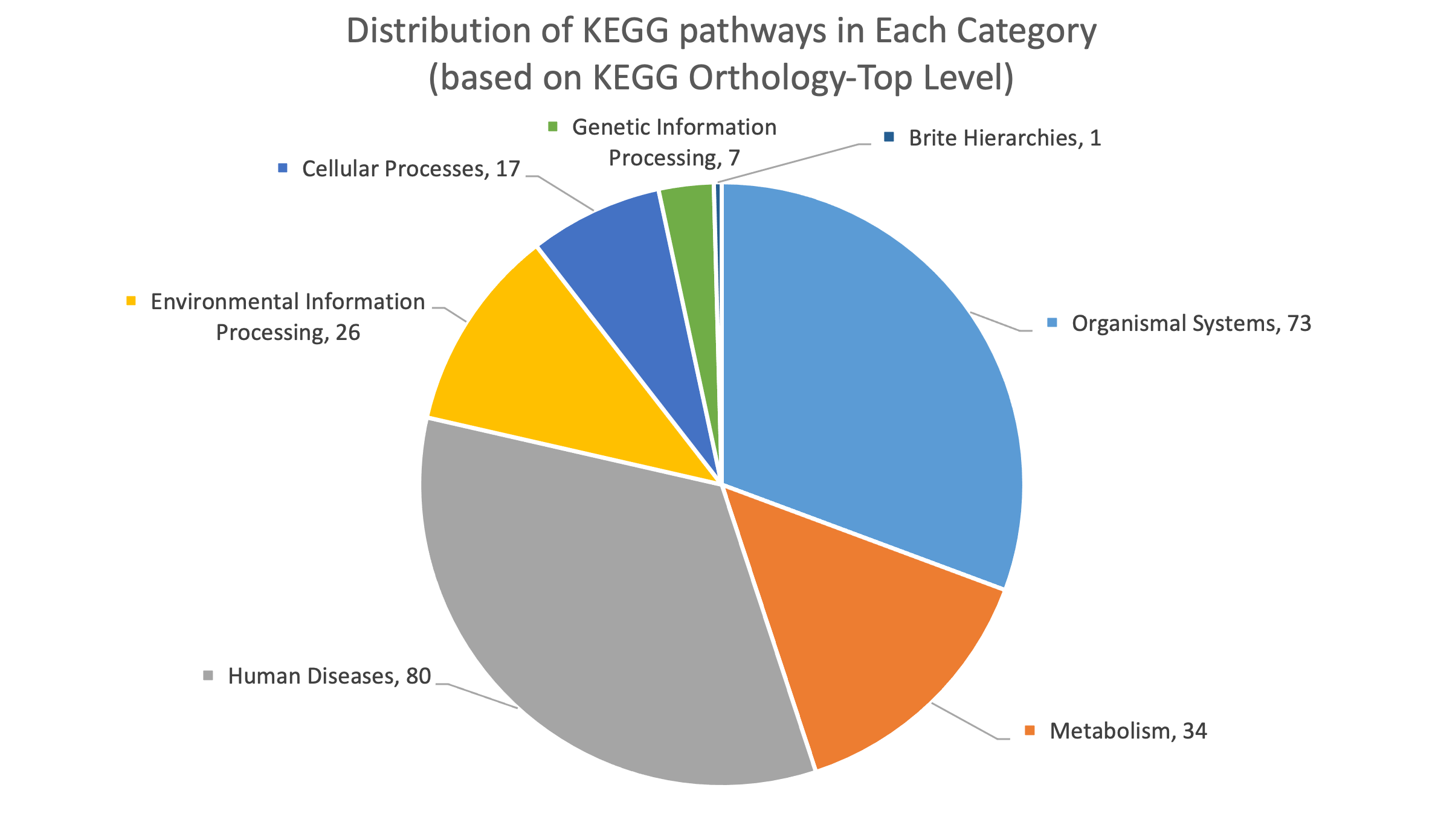
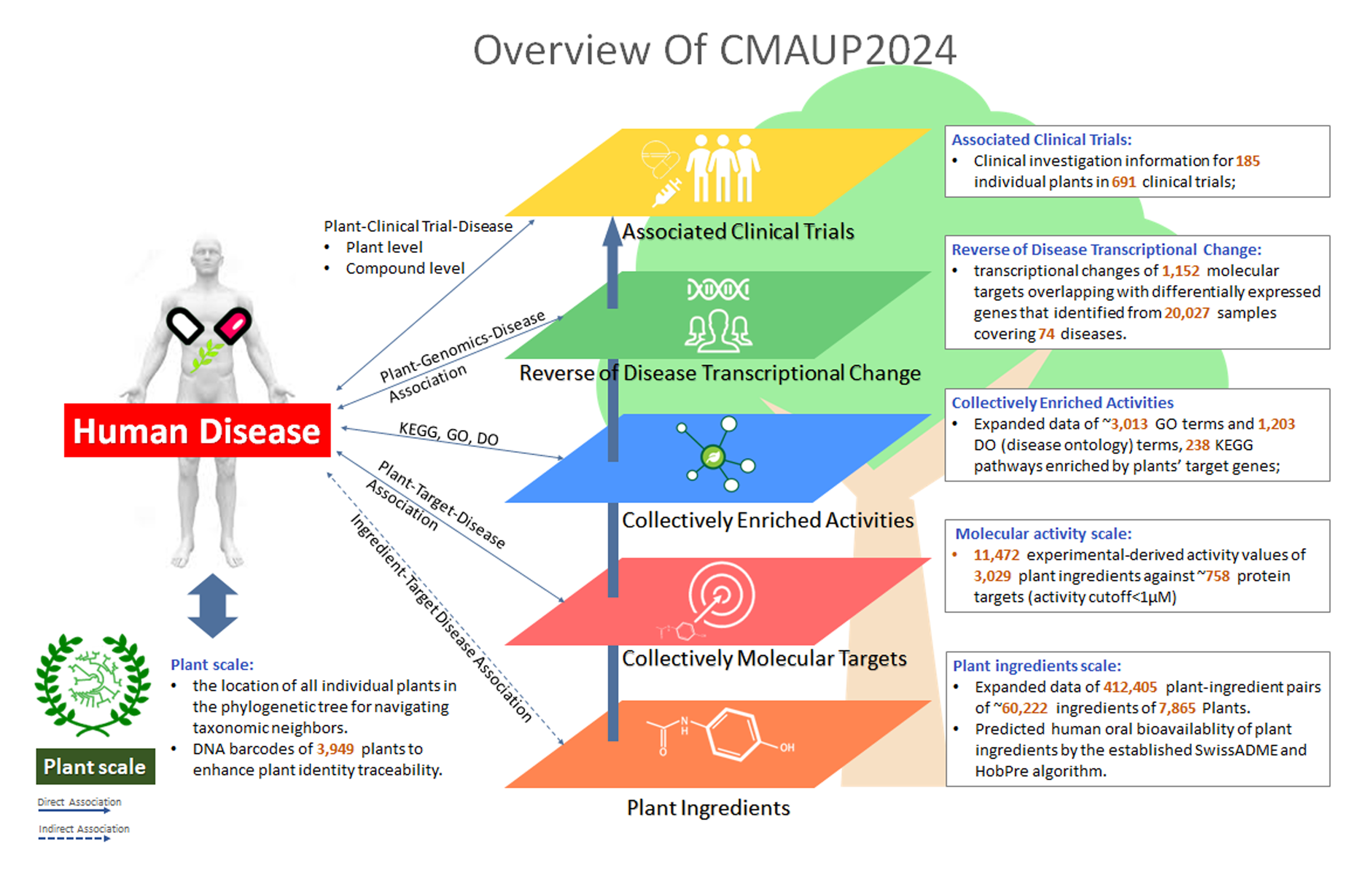
To better support the research community, we significantly updated the CMAUP database by extending the collective molecular activities of palnts to more data layers and more enriched information. The CMAUP ver-2.0 provides collective molecular activities of 7,865 useful plants, including 2,954 medicinal plants used in 79 countries/regions, on 758 human target proteins and 3,013 Gene Ontology, 238 KEGG pathways, and their relations to 1,399 human diseases. This major update includes multi-scales of collective molecular activities of useful plants, as listed below:
❱❱① Firstly, we extended collective molecular activities of individual plants to patients' transcriptomic change levels, by establishing and analyzing disease-specific transcriptomic datasets (which were collected from public-available RNA-seq datasets for diverse diseases from ARCHS4, recount3 and UCSC Xena databases). The collective reversion activities of individual plants were analyzed by comparing plants' target gene lists to up-regulated differential genes in disease tissues (versus normal samples) by RBO algorithm. Currently, CMAUP includes 1,152 molecular targets of 5,765 plants overlapping with differentially expressed genes that identified from 20,027 samples covering 74 diseases.
(1)DEG Analysis
The R package of Combat-seq [Ref: NAR genomics and bioinformatics 2.3 (2020): lqaa078.] was used to remove batch effects. DEGs were identified using R package DESeq2 with the cutoff of | log2(fold change) | > 1.5 and adjusted P-value <0.001.
(2)Rank Biased Overlap (RBO)
We calculate Rank Biased Overlap (RBO) [Ref: Transactions on Information Systems (TOIS) 28.4 (2010): 1-38.] of each plant and disease using the list of up-regulated gene and targets of plant (<1μM). An online calculating tool was also provide for users to calculate the RBOs of their interested diseases using the target list within CMAUP.
❱❱② Clinical investigation information for 185 individual plants in 691 clinical trials;
❱❱③ Drug development information for 4,694 drug-producing plants with metabolites developed into approved or clinical trial drugs;
❱❱④ Plant and human disease associations for 428,737 associations by therapeutic target, 220,935 associations by reversion of transcriptional change in targeted human populations, 764 associations by clinical trials of individual plants, and 154,121 associations by clinical trials of plant ingredients;
❱❱⑤ The location of all individual plants in the phylogenetic tree for navigating taxonomic neighbors.
Phylogenetic tree projection of individual plants at family level to allow the overview of the distribution of bioactive plants from the phylogenetic view. We used phyloT (https://phylot.biobyte.de/) to generate phylogenetic tree for 372 families and labeled other relevant plant species or genera in CMAUP.
❱❱⑥ DNA barcodes of 3,949 plants. We obtained ITS barcode of plants from database PLANiTS [Ref: Database 2020 (2020): baz155.] - ITS, ITS1 and ITS2 reference dataset for subkingdom Viridiplantae - to enhance plant identity traceability.
❱❱⑦ Predicted human oral bioavailablity of plant ingredients by the established SwissADME and HobPre algorithm.
New data of the predicted bioavailability properties to estimate absorbed ingredients of plants in the blood, which are critical for evaluating therapeutic effect or side-effect of herb medicines. SwissADME [Ref: Scientific reports 7.1 (2017): 42717.] and HobPre [Ref: Journal of Cheminformatics 14.1 (2022): 1-10.] were utilized to evaluate human oral bioavailabilty.SwissADME: six descriptors are used by SwissADME to evaluate the oral bioavailability of a natural product:
☑ LIPO(Lipophility): -0.7 < XLOGP3 < +5.0
☑ SIZE: 150g/mol < MW < 500g/mol
☑ POLAR(Polarity): 20Ų < TPSA < 130Ų
☑ INSOLU(Insolubility): -6 < Log S (ESOL) < 0
☑ INSATU(Insaturation): 0.25 < Fraction Csp3 < 1
☑ FLEX(Flexibility): 0 < Num. rotatable bonds < 9
If 6 descriptors of a natural plant satisfies the above rules, it will be labeled high HOB.
HobPre: A natural plant with HobPre score >0.5 is labeled high human oral availability (HOB)
Features
>> Multiple Search/Browse Options
- Search/Browse Plants by Plant Usage Class
- Search/Browse Plants by Target Protein
- Search/Browse Plants by Target Gene Ontology
- Search/Browse Plants by Target KEGG Pathway
- Search/Browse Plants by Related Disease
- Browse Plants by Geographical Location
- Search Plants by Chemical Ingredient
>> Medicinal Plants Used in Various 26 Asian, 19 African, 23 European, 8 North and South American, and 3 Oceania Countries/Regions
>> Collective Molecular Activities of 7,867 Plants Based on Experiment-Derived (not in-silico predicted) Quantitative Activities
>> View Detailed Activity Values Along With Source References
>> Free Download of All Data
External Links to Other Resources
Entries in CMAUP database were linked to various external databases, including:
- Plant databases: NCBI TaxonomyDB, Plants of teh World Online
- Chemical Databases: PubChem, ZINC, ChEMBL
- Activity and Therapeutical related databases: Uniprot, KEGG, QuickGO, ChEMBL, TTD, Disease Ontology
- Source of RNA-seq data: ARCHS4, recount3, UCSC Xena, TCGA
- Literature Resources: PubMed, doi.org
Statistics of Data Content