Collective Molecular Activities of the Plant: Diphylleia Sinensis
Plant ID: NPO26868
Plant Latin Name: Diphylleia Sinensis
Taxonomy Genus: Diphylleia
Taxonomy Family: Berberidaceae
Plant External Links:
NCBI TaxonomyDB:
382113
Plant-of-the-World-Online:
n.a.
Country/Region:
ChinaTraditional Medicine System:
TCMMedicinal Functions:
Antiseptic; Cancer
China
Overview of Ingredients
20 All known Ingredients in Total
Unique ingredients have been isolated from this plant.Plant-Ingredients Associations were manually curated from publications or collected from other databases.
17 Ingredients with Acceptable Bioavailablity
Unique ingredients exhibit acceptable human oral bioavailablity, according to the criteria of SwissADME [PMID: 28256516] and HobPre [PMID: 34991690]. The criteria details:SwissADME: six descriptors are used by SwissADME to evaluate the oral bioavailability of a natural product:
☑ LIPO(Lipophility): -0.7 < XLOGP3 < +5.0
☑ SIZE: 150g/mol < MW < 500g/mol
☑ POLAR(Polarity): 20Ų < TPSA < 130Ų
☑ INSOLU(Insolubility): -6 < Log S (ESOL) < 0
☑ INSATU(Insaturation): 0.25 < Fraction Csp3 < 1
☑ FLEX(Flexibility): 0 < Num. rotatable bonds < 9
If 6 descriptors of a natural plant satisfy the above rules, it will be labeled high HOB.
HobPre: A natural plant ingredient with HobPre score >0.5 is labeled high human oral availability (HOB)
11 Ingredients with experimental-derived Activity
Unique ingredients have activity data available.Ingredient Structrual Cards

Ingredient ID: NPC94759
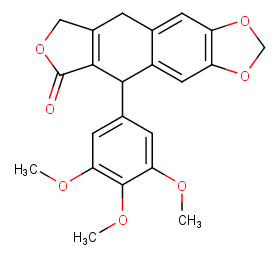
Ingredient ID: NPC40654
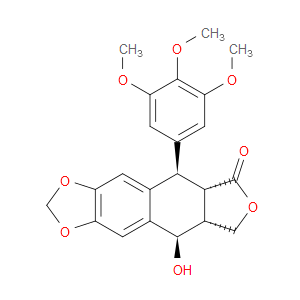
Ingredient ID: NPC32373
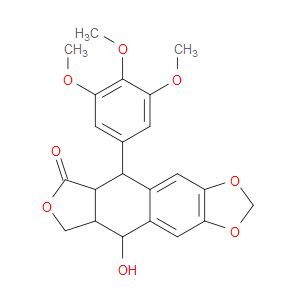
Ingredient ID: NPC318335
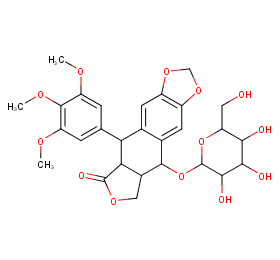
Ingredient ID: NPC301351

Ingredient ID: NPC30009
Ingredient ID: NPC293899
Ingredient ID: NPC284603
Ingredient ID: NPC280403
Ingredient ID: NPC279624

Ingredient ID: NPC262804
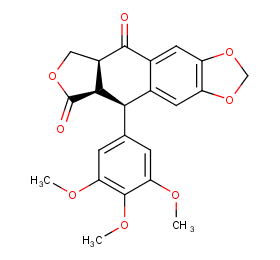
Ingredient ID: NPC239890
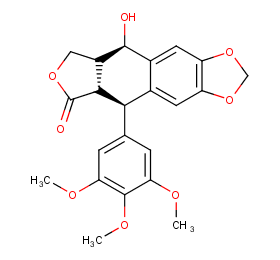
Ingredient ID: NPC226168
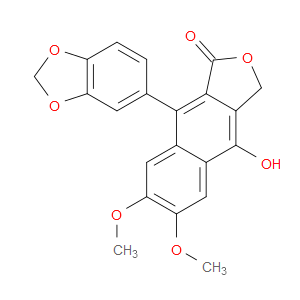
Ingredient ID: NPC22130
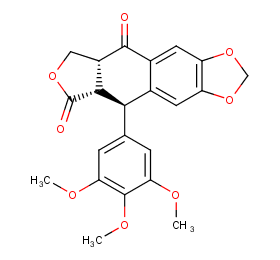
Ingredient ID: NPC209411
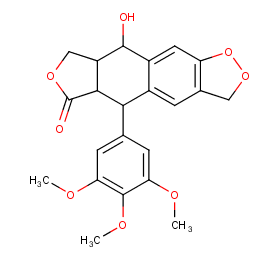
Ingredient ID: NPC194873
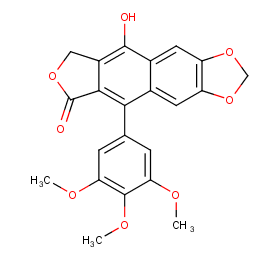
Ingredient ID: NPC174734
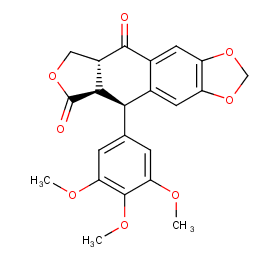
Ingredient ID: NPC169037

Ingredient ID: NPC150943
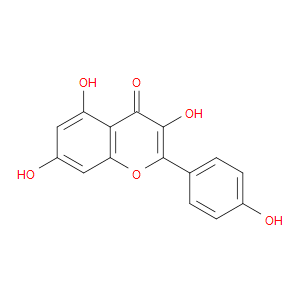
Ingredient ID: NPC116775
Classification of Human Proteins Collectively Targeted by the Plant
Detailed Information of Target Proteins
| Target Type | Protein Class | Gene ID | Protein Name | Uniprot ID | Target ChEMBL ID |
|---|---|---|---|---|---|
| Cytochrome P450 | Cytochrome P450 family 1 | CYP1A2 | Cytochrome P450 1A2 | P05177 | CHEMBL3356 |
| Cytochrome P450 | Cytochrome P450 family 1 | CYP1B1 | Cytochrome P450 1B1 | Q16678 | CHEMBL4878 |
| Cytochrome P450 | Cytochrome P450 family 1 | CYP1A1 | Cytochrome P450 1A1 | P04798 | CHEMBL2231 |
| Therapeutic Target | Lyase | CA12 | Carbonic anhydrase XII | O43570 | CHEMBL3242 |
| Therapeutic Target | Lyase | CA7 | Carbonic anhydrase VII | P43166 | CHEMBL2326 |
| Therapeutic Target | Oxidoreductase | HSD17B2 | Estradiol 17-beta-dehydrogenase 2 | P37059 | CHEMBL2789 |
| Therapeutic Target | Transcription factor | AHR | Aryl hydrocarbon receptor | P35869 | CHEMBL3201 |
Clinical trials associated with plant from natural product (NP) & plant level:
| Clinical trials type | Number of clinical trials | |
|---|---|---|
| 5 | ||
| NCT ID | Title | Condition | Form in clinical use | Associated by plant or compound |
|---|---|---|---|---|
| NCT01725555 | A Study to Assess the Effect of Food on the Bioavailability of the IGF-1R Inhibitor AXL1717 in Patients With Advanced Malignant Tumors | lymphoid neoplasm | Picropodophyllotoxin (NPC32373) | |
| NCT01466647 | A Study of the IGF-1R Inhibitor AXL1717 in Combination With Gemcitabine HCL and Carboplatin to Treat Non-small-cell Lung Cancer (NSCLC) | non-small cell lung carcinoma | Picropodophyllotoxin (NPC32373) | |
| NCT01062620 | A Dose Escalating Clinical Trial of the IGF-1 Receptor Inhibitor AXL1717 in Patients With Advanced Cancer | neoplasm | Picropodophyllotoxin (NPC32373) | |
| NCT01721577 | Phase I/II Trial of AXL1717 in the Treatment of Recurrent Malignant Astrocytomas | anaplastic astrocytoma;anaplastic oligodendroglioma;gliosarcoma | Picropodophyllotoxin (NPC32373) | |
| NCT01561456 | Study of AXL1717 Compared to Docetaxel to Treat Squamous Cell Carcinoma or Adenocarcinoma of the Lung | lung adenocarcinoma;squamous cell carcinoma | Picropodophyllotoxin (NPC32373) |
❱❱❱ Associated Human Diseases and Detailed Association Evidence
How do we define the Plant-Targeted Human Disease Association?
Associated human diseases of an individual plant are summurized based on FOUR types of association evidence, these include:
❶ Association by Therapeutic Target: Bioactive protein targets of the plant were defined in "Molecular Targets" section, target-disease associations collected from TTD database were subsequently used to build the associations between the plant and its targeted human diseases.
❷ Association by Disease Gene Reversion: Plant and a specific disease will be associated when >= 1 plant target gene overlaped with disease's DEGs.
❸ Association by Clinical Trials of Plant: Plant and a specific disease will be associated when >= 1 clinical trial (the plant is the intervetion) can be matched in ClinicalTrials.gov database.
❹ Association by Clinical Trials of Plant Ingredients: Plant and a specific disease will be associated when >= 1 clinical trial (the plant ingredient is the intervetion) can be matched in ClinicalTrials.gov database.
Associated Disease of the Plant |
Association Type & Detailed Evidence |
|---|---|
Mesothelioma of pleuraDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2C26.0 |
CSNK2A2,AHR,CA7,DPP4,CA12,TYR,HSD17B2
|
Serous cystadenoma,borderline malignancy of ovaryDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2C73.4 |
AHR,CSNK2A3,CA12,CYP1A2,CISD1,HSD17B2
|
Adenocarcinoma of pancreasDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2C10.0 |
CA7,DPP4,CISD1,CYP1B1,CYP2C9,HSD17B2
|
Malignant neoplasms of thymusDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2C27 |
DPP4,CISD1,CYP1B1,CYP2C9,HSD17B2
|
Germ cell tumour of testisDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2C80.2 |
AHR,DPP4,CA12,CYP2C9,HSD17B2
|
Carcinosarcoma of uterusDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2C76.43 |
CA7,DPP4,CISD1,HSD17B2
|
Diffuse large B-cell lymphomasDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2A81 |
CSNK2A2,AHR,CA12,CISD1
|
Malignant neoplasms of thyroid gland, unspecifiedDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2D10.Z |
DPP4,CYP1B1,CA12,CA7
|
Glioblastoma of brainDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2A00.00 |
CSNK2A3,DPP4,CA12,CYP1B1
|
Malignant neoplasms of adrenal glandDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2D11 |
HSD17B2,DPP4,CA12,CA7
|
Solid tumour/cancerDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2A00-2F9Z |
AHR,CSNK2A1,CA12
|
Squamous cell carcinoma of bronchus or lungDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2C25.2 |
CA7,CA12,CYP2C9
|
Adenocarcinoma of bronchus or lungDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2C25.0 |
CA7,CA12,CYP2C9
|
Malignant neoplasms of corpus uteri, unspecifiedDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2C76.Z |
CYP2C9,HSD17B2,CYP1A1
|
Renal cell carcinoma, chromophobe typeDisease Category: X.Extension CodesDisease ICD-11 Code: XH6153 |
CA12,CYP1A2,CYP1A1
|
Bacterial infectionDisease Category: 01.Certain infectious or parasitic diseasesDisease ICD-11 Code: 1A00-1C4Z |
CA12,CYP2C9
|
PsoriasisDisease Category: 14.Diseases of the skinDisease ICD-11 Code: EA90 |
AHR,DPP4
|
Osteoarthritis, unspecifiedDisease Category: 15.Diseases of the musculoskeletal system or connective tissueDisease ICD-11 Code: FA0Z |
HSD17B2,CA12
|
Other specified malignant neoplasms of kidney, except renal pelvisDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2C90.Y |
TYR,CYP2C9
|
Other specified malignant neoplasms ofcolonDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2B90.Y |
CYP1A1,TYR
|
Hepatocellular carcinoma of liverDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2C12.02 |
CA12,CA12
|
Urothelial carcinoma of bladderDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2C94.2 |
CYP2C9,CYP1A2
|
Cytomegaloviral diseaseDisease Category: 01.Certain infectious or parasitic diseasesDisease ICD-11 Code: 1D82 |
CYP2C9,CYP1A1
|
Carpal tunnel syndromeDisease Category: 08.Diseases of the nervous systemDisease ICD-11 Code: 8C10.0 |
DPP4,CYP1B1
|
Myelodysplastic syndromeDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2A37 |
AHR
|
Liver cancerDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2C12 |
CSNK2A1
|
Myocardial infarctionDisease Category: 11.Diseases of the circulatory systemDisease ICD-11 Code: BA41-BA43 |
DPP4
|
Seborrhoeic dermatitisDisease Category: 14.Diseases of the skinDisease ICD-11 Code: EA81 |
CA12
|
Acquired hypermelanosisDisease Category: 14.Diseases of the skinDisease ICD-11 Code: ED60 |
TYR
|
Diabetes mellitus, type unspecifiedDisease Category: 05.Endocrine, nutritional or metabolic diseasesDisease ICD-11 Code: 5A14 |
DPP4
|
MelanomaDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2C30 |
TYR
|
Ureteral cancerDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2C92 |
AHR
|
Chronic arterial occlusive diseaseDisease Category: 11.Diseases of the circulatory systemDisease ICD-11 Code: BD4Z |
DPP4
|
Atopic eczemaDisease Category: 14.Diseases of the skinDisease ICD-11 Code: EA80 |
AHR
|
Type 2 diabetes mellitusDisease Category: 05.Endocrine, nutritional or metabolic diseasesDisease ICD-11 Code: 5A11 |
DPP4
|
Coronavirus infectionDisease Category: 01.Certain infectious or parasitic diseasesDisease ICD-11 Code: 1D92 |
CSNK2A1
|
Breast cancerDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2C60-2C6Y |
CA12
|
Metabolic disorderDisease Category: 05.Endocrine, nutritional or metabolic diseasesDisease ICD-11 Code: 5C50-5D2Z |
DPP4
|
ThrombocytopeniaDisease Category: 03.Diseases of the blood or blood-forming organsDisease ICD-11 Code: 3B64 |
AHR
|
Multiple myelomaDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2A83 |
AHR
|
Autism spectrum disorderDisease Category: 06.Mental, behavioural or neurodevelopmental disordersDisease ICD-11 Code: 6A02 |
DPP4
|
Malignant haematopoietic neoplasmDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2B33 |
DPP4
|
Acquired hypomelanotic disorderDisease Category: 14.Diseases of the skinDisease ICD-11 Code: ED63 |
TYR
|
Acute diabete complicationDisease Category: 05.Endocrine, nutritional or metabolic diseasesDisease ICD-11 Code: 5A2Y |
DPP4
|
Oligodendroglioma, anaplasticDisease Category: X.Extension CodesDisease ICD-11 Code: XH8844 |
NCT01721577
|
Squamous cell carcinomaDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2B60-2D01 |
NCT01561456
|
NeoplasmsDisease Category: 02.NeoplasmsDisease ICD-11 Code: 02 |
NCT01062620
|
GliosarcomaDisease Category: X.Extension CodesDisease ICD-11 Code: XH9RC8 |
NCT01721577
|
AdenocarcinomaDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2D40 |
NCT01561456
|
Non-small cell lung cancerDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2C25 |
NCT01466647
|
Neoplasms of haematopoietic or lymphoid tissues, unspecifiedDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2B3Z |
NCT01725555
|
Astrocytoma, anaplasticDisease Category: X.Extension CodesDisease ICD-11 Code: XH96C7 |
NCT01721577
|
Coeliac diseaseDisease Category: 13.Diseases of the digestive systemDisease ICD-11 Code: DA95 |
CYP1A1
|
Myelodysplastic syndromes, unspecifiedDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2A3Z |
CYP2C9
|
Malignant neoplasms of biliary tract, distal bile ductDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2C15 |
CSNK2A3
|
HepatoblastomaDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2C12.01 |
CA12
|
LeishmaniasisDisease Category: 01.Certain infectious or parasitic diseasesDisease ICD-11 Code: 1F54 |
DPP4
|
Atrial fibrillationDisease Category: 11.Diseases of the circulatory systemDisease ICD-11 Code: BC81.3 |
CA7
|
Myotonic dystrophyDisease Category: 08.Diseases of the nervous systemDisease ICD-11 Code: 8C71.0 |
CYP1A1
|
West Nile virus infectionDisease Category: 01.Certain infectious or parasitic diseasesDisease ICD-11 Code: 1D46 |
DPP4
|
Pneumonia due to PseudomonasaeruginosaDisease Category: 12.Diseases of the respiratory systemDisease ICD-11 Code: CA40.05 |
DPP4
|
Other specified malignant neoplasms of bronchus or lungDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2C25.Y |
CA12
|
Superficial ovarian endometriosisDisease Category: 16.Diseases of the genitourinary systemDisease ICD-11 Code: GA10.B4 |
DPP4
|
Chronic rhinosinusitisDisease Category: 12.Diseases of the respiratory systemDisease ICD-11 Code: CA0A |
CYP1A1
|
Systemic lupus erythematosusDisease Category: 04.Diseases of the immune systemDisease ICD-11 Code: 4A40.0 |
CYP1A1
|
Invasive carcinoma of breastDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2C61 |
CA12
|
Heart failure, unspecifiedDisease Category: 11.Diseases of the circulatory systemDisease ICD-11 Code: BD1Z |
CYP1B1
|
Ebola virus diseaseDisease Category: 01.Certain infectious or parasitic diseasesDisease ICD-11 Code: 1D60.01 |
HSD17B2
|
Gliomas of brainDisease Category: 02.NeoplasmsDisease ICD-11 Code: 2A00.0 |
AHR
|

